Mbj-0110, a novel cyclopeptide isolated from the fungus penicillium sp. F25267
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

We have constructed an isolated natural compound library designed to facilitate extensive biological screenings.1 The library consists of over 1000 isolates, including over 140 ‘JBIR
compounds’ that were discovered in our laboratory. To enrich this library, we recently initiated a screening program for rare microbial products using the advanced compound-identification
system designated as ‘MBJ’s special selection’.2, 3 As a result, our program yielded novel compounds named ‘MBJ compounds’, such as a cytotoxic hydroxamate MBJ-0003 from _Micromonospora_ sp.
29867;4 cytotoxic eremophilane derivatives MBJ-0009 and MBJ-0010 from _Nectria_ sp. f26111;5 MBJ-0011, MBJ-0012 and MBJ-0013 from _Apiognomonia_ sp. f24023;2 cytotoxic chaetoglobosin
derivatives MBJ-0038, MBJ-0039 and MBJ-0040 from _Chaetomium_ sp. f24230;3 bicyclic depsipeptides MBJ-0086 and MBJ-0087 from _Sphaerisporangium_ sp. 33226;6 and aziridine-containing peptide
MBJ-0035 from _Streptosporangium_ sp. 32552.7 Further screening for novel compounds led to the identification of MBJ-0110 (1) from the culture of _Penicillium_ sp. f25267. Herein we report
the fermentation, isolation, structure elucidation and preliminary biological activity data. _Penicillium_ sp. f25267 was isolated from a soil sample collected in the Shiga Prefecture,
Japan. The strain was cultured in 250-ml Erlenmeyer flasks, each containing 25 ml of a seed medium consisting of 2% potato starch (Tobu Tokachi Nosan Kako Agricultural Cooperative Assoc.,
Hokkaido, Japan), 1% glucose (Junsei Chemical, Tokyo, Japan), 2% soybean powder (SoyPro, J-Oil Mills, Tokyo, Japan), 0.1% KH2PO4 and 0.05% MgSO4·7H2O (pH 7.4 before sterilization). The
flasks were incubated on a rotary shaker (220 r.p.m.) at 25 °C for 3 days. Aliquots (0.5 ml) of the broth were transferred to 500-ml Erlenmeyer flasks containing 50 ml of a production medium
of the same composition, which were then cultured on a rotary shaker (220 r.p.m.) at 25 °C for 4 days. The whole culture broth (2 l) was extracted with an equal volume of _n_-BuOH. After
concentration _in vacuo_, the extract was successively partitioned between EtOAc (350 ml × 3) and H2O (300 ml). The aqueous layer was evaporated to dryness and the residue (1.4 g) was
fractionated by reversed-phase medium-pressure liquid chromatography (Purif-Pack ODS-30, Shoko Scientific, Yokohama, Japan; 40–100% aq. MeOH with 10% stepwise increments in the MeOH
concentration). The fractions were monitored using an ultra performance liquid chromatography-diode array detection-evaporative light scattering-mass spectrometry system and 1 was isolated
based on peak-guided fractionation. The 50% MeOH eluate (30.9 mg) was subjected to preparative reversed-phase HPLC using a Capcell Pak C18 MG II column (20 mm inside diameter (i.d.) × 150
mm; Shiseido, Tokyo, Japan) with a solvent system of 20% CH3CN/H2O containing 0.1% formic acid (flow rate: 10 ml min–1), to yield semi-purified 1 (6.9 mg, retention time (Rt)=15.3 min).
Final purification was carried out by preparative HPLC using an X-Bridge C18 column (19 mm i.d. × 150 mm; Waters, Milford, MA, USA) with a solvent system of 20% CH3CN/H2O containing 0.1%
formic acid (flow rate: 10 ml min–1) to afford 1 (3.5 mg, Rt=13.3 min). MBJ-0110 (1) was obtained as a colorless amorphous powder: [α]24D –186 (MeOH; _c_ 0.18); UV end; IR (attenuated total
reflectance) _ν_max: 3400 (hydroxy) and 1683 (carbonyl) cm−1. The molecular formula of 1 was established as C27H41N5O8 by high-resolution (HR)-ESI–MS (_m/z_ 564.3018 [M+H]+, calcd for
C27H42N5O8 _m/z_ 564.3033). Its peptidic nature was evident from the resonances corresponding to α-methine protons (_δ_H 4.00–5.08) and the resonances corresponding to the carbonyl carbons
(_δ_C 169.0–175.9) in the 1H and 13C NMR spectra of 1, respectively. The direct connectivity between protons and carbons was established by a HSQC spectrum; Table 1 summarizes the 13C and 1H
NMR spectroscopic data for 1. The 1H sequences and 1H−13C long-range couplings from α-methine protons to the corresponding amide carbonyl carbons, which were elucidated by double
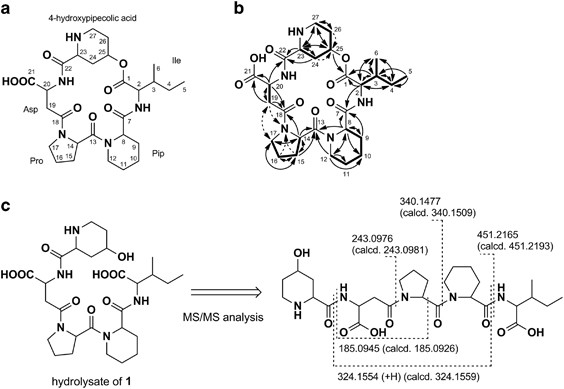
quantum-filtered COSY and constant time-HMBC8 spectra, respectively, revealed the involvement of an isoleucine (Ile), a pipecolic acid (Pip), a proline (Pro) and an aspartic acid (Asp)
residue, as shown in Figure 1b. In addition to the above-mentioned amino-acid moieties, the presence of a 4-hydroxypipecolic acid (C-22 to C-27) moiety was proved based on a 1H sequence from
an α-methine proton H-23 (_δ_H 4.00) to nitrogen-bearing methylene protons H2-27 (_δ_H 3.39 and 2.98) via aliphatic methylene protons H2-24 (_δ_H 2.62 and 2.07), an oxymethine proton H-25
(_δ_H 5.17, _δ_C 68.9) and aliphatic protons H2-26 (_δ_H 2.19, 2.02), and 1H–13C long-range couplings from H-23 and H-24 (_δ_H 2.07) to an amide carbonyl carbon C-22 (_δ_C 169.0), and from
H2-27 to an α-methine carbon C-23 (_δ_C 55.5). The amino-acid sequence in 1 was determined by the HMBC correlations from an α-methine proton H-2 (_δ_H 4.36) to a carbonyl carbon C-7 (_δ_C
173.0), from an α-methine proton H-8 (_δ_H 5.08) and ɛ-methylene protons H2-12 (_δ_H 3.97 and 3.00) to a carbonyl carbon C-13 (_δ_C 175.9), from an α-methine proton H-14 (_δ_H 4.95) to a
carbonyl carbon C-18 (_δ_C 171.3), from an α-methine proton H-20 (_δ_H 4.82) to C-22 and from H-25 to an ester carbonyl carbon C-1 (_δ_C 169.7). Although only 1H–13C HMBC information
suggested two structural possibilities, α- and β-aspartyl amide linkages, we concluded that the aspartyl acid moiety is linked to adjacent Pro by β-amino bond because of the existence of a
ROESY correlation between Hb-17 (_δ_H 3.70) and Ha-19 (_δ_H 3.10) and 1H–15N HMBC correlations from H2-16 (_δ_H 2.08, 1.96), Hb-15 (_δ_H 1.80) and Hb-19 (_δ_H 2.96) to a nitrogen atom of Pro
(_δ_N 140). Therefore, the structure of 1 was determined as shown in Figure 1b. To verify the proposed structure, 1 was treated with 0.1 N NaOH overnight at room temperature, followed by
ESI–MS/MS analysis of the alkaline hydrolysate (molecular formula: C27H43N5O9; HR-ESI–MS: [M+H]+ _m/z_ 582.3159, C27H44N5O9 582.3139). The ESI–MS/MS data showed major fragment ions (_m/z_
185.0945, 243.0976, 324.1554, 340.1477 and 451.2165) that supported the proposed structure (Figure 1c). The multiplicity and a large 1H spin coupling constant value of H-23 (doublet,
_J_H–H=7.2 Hz) and ROESY correlations between H-23/Hax-27 (_δ_H 2.98) and H-23/Heq-24 (_δ_H 2.62) implied that the piperidine ring is in the chair conformation and the H-23 is axially
orientated. In addition, the broad singlet signal of H-25 proved its equatorial orientation. Taken together, the relative configurations of C-23 and C-25 were determined as 23_S_* and
25_S_*, respectively. The absolute configurations of the amino-acid residues were determined to be l-Pro, l-Pip and l-Asp by using Marfey’s method.9 A portion of 1 (0.4 mg) was hydrolyzed in
6 n HCl at 110 °C for 12 h and then dried under air flow. The resulting hydrolysate was treated with 0.1 m NaHCO3 (200 μl) and 1% _N_-(5-fluoro-2,4-dinitrophenyl)-l-alaninamide (l-FDAA) in
Me2CO (100 μl) at 40 °C for 30 min. Amino-acid standards were derivatized with l-FDAA in a similar manner. The Marfey’s derivatives were analyzed using a HPLC–MS system as follows: a Capcell
Pak C18 MG II column (4.6 mmi.d. × 150 mm) was developed with a linear gradient system of water/MeCN with 0.1% formic acid (20–50% MeCN, 15 min; flow rate, 1.0 ml min–1). FDAA derivatives
were detected by absorption at 340 nm, and assignment was secured by ion-selective monitoring. The retention times of the standard FDAA derivatives were as follows: l-Asp, 7.9 min; d-Asp,
8.2 min; l-Pip, 13.6 min; d-Pip, 12.8 min; l-Pro, 10.1 min; d-Pro, 10.7 min; l-Ile, 14.7 min; d-Ile, 16.8 min; l-_allo_-Ile, 14.7 min; and d-_allo_-Ile, 16.7 min. The retention times of the
FDAA derivatives of 1 were as follows: Asp, 7.9 min; Pip, 13.6 min; Pro, 10.1 min; and Ile, 14.7 min. The absolute configuration of the Ile residue in 1 was established by HPLC comparison of
the 2,3,4,6-tetra-_O_-acetyl-β-d-glucopyranosyl isothiocyanate (GITC) derivative of hydrolysate of 1 with standard samples.10 Triethylamine (50 μl) and a GITC solution (250 μl, prepared at
3.9 mg ml–1 in CH3CN) were added to the acid hydrolysate of 1 or an authentic amino-acid standard. The reaction mixture was kept at room temperature for 30 min and the reaction was then
quenched by adding 40 μl of MeCN–5% AcOH in H2O (1:1). Analysis of the GITC derivatives was performed on a Capcell Pak ADME column (4.6 mm i.d. × 150 mm; Shiseido) employing an isocratic
elution of 40% CH3CN containing 0.1% formic acid (1.0 ml min–1). GITC derivatives were detected by absorption at 248 nm, and assigned by ion-selective monitoring. The retention times of the
GITC derivatives were as follows: l-Ile, 11.6 min and l-_allo_-Ile, 11.3 min. The retention time (11.6 min) of the GITC derivative of 1 implied that the Ile residue in 1 is L-Ile. We
evaluated the cytotoxic and antimicrobial activities of 1, but it showed neither cytotoxicity to human ovarian adenocarcinoma SKOV-3 cell lines (IC50>100 μM) or human malignant pleural
mesothelioma ACC-MESO-1 cell lines (IC50>100 μM), nor antimicrobial activity against _Micrococcus luteus_ and _Bacillus subtilis_. The obtained structure of 1 is very rare in nature; only
petrosifungins A and B,11 and JBIR-113, -114 and -11512 have been isolated as pipecolic acid-containing peptides of fungal origin. To the best of our knowledge, there are no reports in the
literature of peptide compounds possessing the 4-hydroxypipecolic acid moiety. REFERENCES * Kawahara, T., Nagai, A., Takagi, M. & Shin-ya, K. JBIR-137 and JBIR-138, new secondary
metabolites from _Aspergillus_ sp. fA75. _J. Antibiot_. 65, 535–538 (2012). Article CAS Google Scholar * Kawahara, T. _et al_. Three eremophilane derivatives, MBJ-0011, MBJ-0012 and
MBJ-0013, from an endophytic fungus _Apiognomonia_ sp. f24023. _J. Antibiot_. 66, 299–302 (2013). Article CAS Google Scholar * Kawahara, T. _et al_. New chaetoglobosin derivatives,
MBJ-0038, MBJ-0039 and MBJ-0040, isolated from the fungus _Chaetomium_ sp. f24230. _J. Antibiot_. 66, 727–730 (2013). Article CAS Google Scholar * Kawahara, T. _et al_. New hydroxamate
metabolite, MBJ-0003, from _Micromonospora_ sp. 29867. _J. Antibiot_. 67, 261–263 (2014). Article CAS Google Scholar * Kawahara, T. _et al_. Cytotoxic sesquiterpenoids MBJ-0009 and
MBJ-0010 from a saprobic fungus _Nectria_ sp. f26111. _J. Antibiot_. 66, 567–569 (2013). Article CAS Google Scholar * Kawahara, T. _et al_. MBJ-0086 and MBJ-0087, new bicyclic
depsipeptides from _Sphaerisporangium_ sp. 33226. _J. Antibiot_. 68, 67–70 (2015). Article CAS Google Scholar * Kawahara, T. _et al_. MBJ-0034 and MBJ-0035, new aziridine-containing
peptides from _Streptosporangium_ sp. 32552. _J. Antibiot_. 67, 577–580 (2014). Article CAS Google Scholar * Furihata, K. & Seto, H. Constant time HMBC (CT-HMBC), a new HMBC technique
useful for improving separation of cross peaks. _Tetrahedron Lett_. 39, 7337–7340 (1998). Article CAS Google Scholar * Marfey, P. Determination of D-amino acids. II. Use of a
bifunctional reagent, 1,5- difluoro-2,4-dinitrobenzene. _Carlsberg Res. Commun_. 49, 591–596 (1984). Article CAS Google Scholar * Nimura, N., Ogura, H. & Kinoshita, T. Reversed-phase
liquid chromatographic resolution of amino acid enantiomers by derivatization with 2,3,4,5-tetra-_O_-acetyl-β-D-glucopyranosyl isothiocyanate. _J. Chromatogr_. 202, 375–379 (1980). Article
CAS Google Scholar * Bringmann, G., Lang, G., Steffens, S. & Schaumann, K. Petrosifungins A and B, novel cyclodepsipeptides from a sponge-derived strain of _Penicillium
brevicompactum_. _J. Nat. Prod_. 67, 311–315 (2004). Article CAS Google Scholar * Kawahara, T., Takagi, M. & Shin-ya, K. Three new depsipeptides, JBIR-113, JBIR-114 and JBIR-115,
isolated from a marine sponge-derived _Penicillium_ sp. fS36. _J. Antibiot_. 65, 147–150 (2012). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported in
part by a grant ‘Project focused on developing key technologies for discovering and manufacturing drugs for next-generation treatment and diagnosis’ from the Ministry of Economy, Trade and
Industry (METI). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Japan Biological Informatics Consortium (JBIC), Koto-ku, Tokyo, Japan Teppei Kawahara, Ikuko Kozone & Miho Izumikawa *
Bioresource Laboratories, MicroBiopharm Japan Co., Ltd. (MBJ), Iwata, Shizuoka, Japan Masashi Itoh, Noriaki Sakata & Toshio Tsuchida * National Institute of Advanced Industrial Science
and Technology (AIST), Koto-ku, Tokyo, Japan Kazuo Shin-ya Authors * Teppei Kawahara View author publications You can also search for this author inPubMed Google Scholar * Masashi Itoh View
author publications You can also search for this author inPubMed Google Scholar * Ikuko Kozone View author publications You can also search for this author inPubMed Google Scholar * Miho
Izumikawa View author publications You can also search for this author inPubMed Google Scholar * Noriaki Sakata View author publications You can also search for this author inPubMed Google
Scholar * Toshio Tsuchida View author publications You can also search for this author inPubMed Google Scholar * Kazuo Shin-ya View author publications You can also search for this author
inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Kazuo Shin-ya. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kawahara, T., Itoh, M., Kozone, I. _et al._ MBJ-0110, a novel cyclopeptide isolated from the fungus _Penicillium_ sp. f25267. _J
Antibiot_ 69, 66–68 (2016). https://doi.org/10.1038/ja.2015.78 Download citation * Received: 07 April 2015 * Revised: 08 June 2015 * Accepted: 15 June 2015 * Published: 08 July 2015 * Issue
Date: January 2016 * DOI: https://doi.org/10.1038/ja.2015.78 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative