Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

The architecture of human chromosomes in interphase nuclei is still largely unknown. Microscopy studies have indicated that specific regions of chromosomes are located in close proximity to
the nuclear lamina (NL)1,2,3. This has led to the idea that certain genomic elements may be attached to the NL, which may contribute to the spatial organization of chromosomes inside the
nucleus. However, sequences in the human genome that interact with the NL in vivo have not been identified. Here we construct a high-resolution map of the interaction sites of the entire
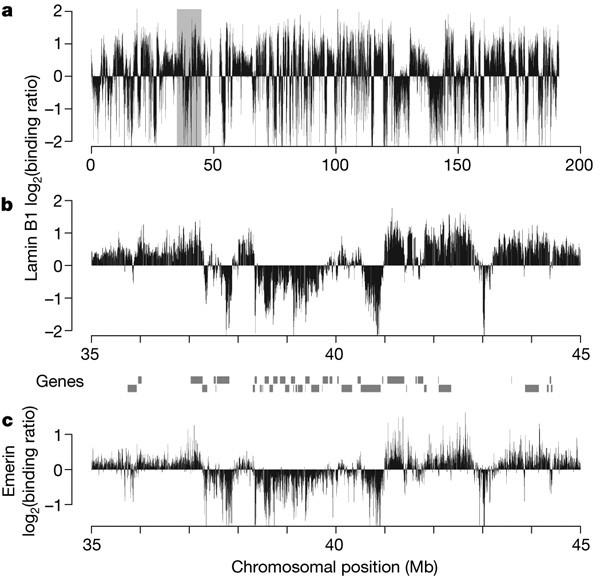
genome with NL components in human fibroblasts. This map shows that genome–lamina interactions occur through more than 1,300 sharply defined large domains 0.1–10 megabases in size. These
lamina-associated domains (LADs) are typified by low gene-expression levels, indicating that LADs represent a repressive chromatin environment. The borders of LADs are demarcated by the
insulator protein CTCF, by promoters that are oriented away from LADs, or by CpG islands, suggesting possible mechanisms of LAD confinement. Taken together, these results demonstrate that
the human genome is divided into large, discrete domains that are units of chromosome organization within the nucleus.
Microarray probe coordinates and data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE8854.
Nature 453, 948–951 (2008); doi: 10.1038/nature06947 In this Letter, Fig. 4g and Supplementary Fig. 5f depict an analysis of H3K9me3 (not H3K9me2) ChIP data. In addition, three sentences in
the third paragraph of the ‘Microarray data analysis’ section of the Methods should read: “Gene expression data and H3K27me3 ChIP data are from Tig3 lung fibroblasts18.
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007)
We thank R. Versteeg, M. Indemans, R. van Driel and O. Giromus for help with FISH; L. Oomen for help with confocal microscopy; A. Bracken and K. Blahnik for data sets; the NKI microarray
facility for technical support; E. de Wit for preliminary data analysis; and M. Fornerod, F. van Leeuwen, H.J. Bussemaker, M. van Lohuizen and members of the B.v.S. laboratory for helpful
suggestions. This study was supported by the Netherlands Cancer Society, the Netherlands Genomics Initiative, and an European Young Investigator Award to B.v.S. E.B. was supported by NWO
Rubicon.
Author Contributions L.G. performed DamID and related experiments; M.B.F. and L.G. performed immunofluorescence microscopy studies; E.B. conducted FISH experiments in the W.d.L. laboratory,
with technical support from L.G., B.H.E., A.d.K. and W.T.; L.P. designed microarrays; L.P. and B.v.S. performed data analysis together with L.G.; W.M. and L.W. developed motif and repeat
search algorithms. L.G. and B.v.S. conceived and designed the study and wrote the manuscript.
Division of Molecular Biology, Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands,
Lars Guelen, Ludo Pagie, Wouter Meuleman, Marius B. Faza, Wendy Talhout, Lodewyk Wessels & Bas van Steensel
Department of Clinical Genetics, Erasmus Medical Centre, PO Box 2040, 3000 CA Rotterdam, The Netherlands,
Faculty of Electrical Engineering, Mathematics and Computer Science, Delft University of Technology, Mekelweg 4, 2628 CD Delft, The Netherlands ,
The file contains Supplementary Figures 1-8 and Legends. This file contains a cartoon model summarizing the main results (Supplementary Figure 1) and seven figures showing supporting data
and analyses (Supplementary Figures 2-8). (PDF 5445 kb)
The file contains Supplementary Data. This file is a flat text file in GFF format (http://www.sanger.ac.uk/Software/formats/GFF/) listing the positions of all 1,344 LADs (NCBI build 36).
Score (column 6) indicates the fraction of array probes inside the LAD with a positive Lamin B1 DamID logratio, after applying a running median filter with window size 5. (TXT 119 kb)
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
The architecture of human chromosomes within the nucleus is still a mystery. The nuclear lamina is thought to anchor specific DNA elements to help organize the genome within the nucleus. Van
Steensel and colleagues report a high-resolution map of the sites of the human genome that contact the nuclear lamina. The map shows that interactions with the lamina occur via large yet
sharply demarcated genomic domains. This domain structure is linked to the epigenetic and transcriptional profile of the genome. Many domain borders are marked by specific sequence elements,
indicating that the higher-order folding of chromosomes is in part encoded in the genome itself.