A fast, robust and tunable synthetic gene oscillator
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT One defining goal of synthetic biology is the development of engineering-based approaches that enable the construction of gene-regulatory networks according to ‘design
specifications’ generated from computational modelling1,2,3,4,5,6. This approach provides a systematic framework for exploring how a given regulatory network generates a particular
phenotypic behaviour. Several fundamental gene circuits have been developed using this approach, including toggle switches7 and oscillators8,9,10, and these have been applied in new contexts
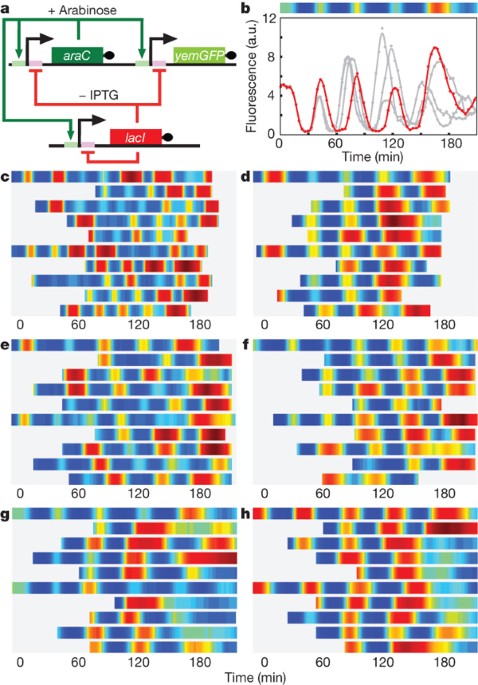
such as triggered biofilm development11 and cellular population control12. Here we describe an engineered genetic oscillator in _Escherichia coli_ that is fast, robust and persistent, with
tunable oscillatory periods as fast as 13 min. The oscillator was designed using a previously modelled network architecture comprising linked positive and negative feedback loops1,13. Using
a microfluidic platform tailored for single-cell microscopy, we precisely control environmental conditions and monitor oscillations in individual cells through multiple cycles. Experiments
reveal remarkable robustness and persistence of oscillations in the designed circuit; almost every cell exhibited large-amplitude fluorescence oscillations throughout observation runs. The
oscillatory period can be tuned by altering inducer levels, temperature and the media source. Computational modelling demonstrates that the key design principle for constructing a robust
oscillator is a time delay in the negative feedback loop, which can mechanistically arise from the cascade of cellular processes involved in forming a functional transcription factor. The
positive feedback loop increases the robustness of the oscillations and allows for greater tunability. Examination of our refined model suggested the existence of a simplified oscillator
design without positive feedback, and we construct an oscillator strain confirming this computational prediction. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only
$3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A SYNTHETIC POPULATION-LEVEL
OSCILLATOR IN NON-MICROFLUIDIC ENVIRONMENTS Article Open access 13 May 2023 FROM RESONANCE TO CHAOS BY MODULATING SPATIOTEMPORAL PATTERNS THROUGH A SYNTHETIC OPTOGENETIC OSCILLATOR Article
Open access 23 August 2024 INDEPENDENT CONTROL OF AMPLITUDE AND PERIOD IN A SYNTHETIC OSCILLATOR CIRCUIT WITH MODIFIED REPRESSILATOR Article Open access 11 January 2022 REFERENCES * Hasty,
J., Dolnik, M., Rottschäfer, V. & Collins, J. J. Synthetic gene network for entraining and amplifying cellular oscillations. _Phys. Rev. Lett._ 88, 148101 (2002) Article ADS Google
Scholar * Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. _Nature_ 420, 224–230 (2002) Article ADS CAS Google Scholar * Tyson, J., Chen, K. & Novak, B.
Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. _Curr. Opin. Cell Biol._ 15, 221–231 (2003) Article CAS Google Scholar * Sprinzak, D.
& Elowitz, M. B. Reconstruction of genetic circuits. _Nature_ 438, 443–448 (2005) Article ADS CAS Google Scholar * Endy, D. Foundations for engineering biology. _Nature_ 438, 449–453
(2005) Article ADS CAS Google Scholar * Andrianantoandro, E., Basu, S., Karig, D. K. & Weiss, R. Synthetic biology: new engineering rules for an emerging discipline. _Mol. Syst.
Biol._ 2, 2006.0028 (2006) Article Google Scholar * Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in _Escherichia coli_ . _Nature_ 403, 339–342
(2000) Article ADS CAS Google Scholar * Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. _Nature_ 403, 335–338 (2000) Article ADS CAS
Google Scholar * Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in _Escherichia coli_ .
_Cell_ 113, 597–607 (2003) Article CAS Google Scholar * Fung, E. et al. A synthetic gene-metabolic oscillator. _Nature_ 435, 118–122 (2005) Article ADS CAS Google Scholar * Kobayashi,
H. et al. Programmable cells: interfacing natural and engineered gene networks. _Proc. Natl Acad. Sci. USA_ 101, 8414–8419 (2004) Article ADS CAS Google Scholar * You, L., Cox, R. S.,
Weiss, R. & Arnold, F. H. Programmed population control by cell-cell communication and regulated killing. _Nature_ 428, 868–871 (2004) Article ADS CAS Google Scholar * Barkai, N.
& Leiber, S. Circadian clocks limited by noise. _Nature_ 403, 267–268 (2000) Article ADS CAS Google Scholar * Lutz, R. & Bujard, H. Independent and tight regulation of
transcriptional units in _Escherichia coli_ via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. _Nucleic Acids Res._ 25, 1203–1210 (1997) Article CAS Google Scholar * Lee, S.
K. et al. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. _Appl. Environ. Microbiol._ 73, 5711–5715 (2007) Article CAS Google Scholar
* Bliss, R. D., Painter, P. R. & Marr, A. G. Role of feedback inhibition in stabilizing the classical operon. _J. Theor. Biol._ 97, 177–193 (1982) Article CAS Google Scholar *
Bratsun, D., Volfson, D., Tsimring, L. S. & Hasty, J. Delay-induced stochastic oscillations in gene regulation. _Proc. Natl Acad. Sci. USA_ 102, 14593–14598 (2005) Article ADS CAS
Google Scholar * Rateitschak, K. & Wolkenhauer, O. Intracellular delay limits cyclic changes in gene expression. _Math. Biosci._ 205, 163–179 (2007) Article MathSciNet CAS Google
Scholar * Mackey, M. & Glass, L. Oscillation and chaos in physiological control systems. _Science_ 197, 287–289 (1977) Article ADS CAS Google Scholar * Jaeger, J. & Reinitz, J.
On the dynamic nature of positional information. _Bioessays_ 28, 1102–1111 (2006) Article CAS Google Scholar * Faith, J. et al. Large-scale mapping and validation of _Escherichia coli_
transcriptional regulation from a compendium of expression profiles. _PLoS Biol._ 5, e8 (2007) Article Google Scholar * Andersen, J. B. et al. New unstable variants of green fluorescent
protein for studies of transient gene expression in bacteria. _Appl. Environ. Microbiol._ 64, 2240–2246 (1998) CAS PubMed PubMed Central Google Scholar * Cookson, S., Ostroff, N., Pang,
W. L., Volfson, D. & Hasty, J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. _Mol. Syst. Biol._ 1, 2005.0024 (2005) Article Google Scholar * Gillespie,
D. T. Exact stochastic simulation of coupled chemical-reactions. _J. Phys. Chem._ 81, 2340–2361 (1977) Article CAS Google Scholar * Hasty, J. et al. Computational studies of gene
regulatory networks: in numero molecular biology. _Nature Rev. Genet._ 2, 268–279 (2001) Article CAS Google Scholar * Ozbudak, E., Thattai, M., Lim, H., Shraiman, B. & van
Oudenaarden, A. Multistability in the lactose utilization network of _Escherichia coli_ . _Nature_ 427, 737–740 (2004) Article ADS CAS Google Scholar * Wang, X., Hao, N., Dohlman, H.
& Elston, T. Bistability, stochasticity, and oscillations in the mitogen-activated protein kinase cascade. _Biophys. J._ 90, 1961–1978 (2006) Article ADS CAS Google Scholar *
Bennett, M. et al. Metabolic gene regulation in a dynamically changing environment. _Nature_ 454, 1119–1122 (2008) Article ADS CAS Google Scholar * Gerland, U., Moroz, J. & Hwa, T.
Physical constraints and functional characteristics of transcription factor-DNA interaction. _Proc. Natl Acad. Sci. USA_ 99, 12015–12020 (2002) Article ADS CAS Google Scholar * Groisman,
A. et al. A microfluidic chemostat for experiments with bacterial and yeast cells. _Nature Methods_ 2, 685–689 (2005) Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We
thank H. Bujard, C. Yang, and Z. Zhang for gifts of reagents, and D. Volfson and M. Simpson for discussions. This work was supported by grants from the National Institutes of Health
(GM69811-01) and the US Department of Defense. AUTHOR CONTRIBUTIONS J.S. and J.H. designed the oscillator circuits, and J.S. constructed the circuits. S.C. performed the microscopy
experiments, and J.S. and S.C. performed the flow cytometry experiments. S.C., L.S.T. and J.H. performed the single-cell data analysis. M.R.B., W.H.M. and L.S.T. performed the computational
modelling. All authors wrote the manuscript. AUTHOR INFORMATION Author notes * Jesse Stricker, Scott Cookson and Matthew R. Bennett: These authors contributed equally to this work. AUTHORS
AND AFFILIATIONS * Department of Bioengineering, University of California, San Diego, La Jolla, California 92093, USA, Jesse Stricker, Scott Cookson, Matthew R. Bennett, William H. Mather
& Jeff Hasty * Institute for Nonlinear Science, University of California, San Diego, La Jolla, California 92093, USA , Matthew R. Bennett, Lev S. Tsimring & Jeff Hasty Authors *
Jesse Stricker View author publications You can also search for this author inPubMed Google Scholar * Scott Cookson View author publications You can also search for this author inPubMed
Google Scholar * Matthew R. Bennett View author publications You can also search for this author inPubMed Google Scholar * William H. Mather View author publications You can also search for
this author inPubMed Google Scholar * Lev S. Tsimring View author publications You can also search for this author inPubMed Google Scholar * Jeff Hasty View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Jeff Hasty. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION This file contains Supplementary
Materials and Methods, Supplementary Figures 1-22 and Supplementary References (PDF 9422 kb) SUPPLEMENTARY MOVIE 1 Supplementary Movie file 1 shows a timelapse microscopy of JS011 cells
continuously induced with 0.7% arabinose and 2 mM IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green. Total time of movie is 228 min with a sampling
rate of one image every 3 min. (MOV 1787 kb) SUPPLEMENTARY MOVIE 2 Supplementary Movie file 2 shows a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 0 mM
IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green. Total time of movie is 219 min with a sampling rate of one image every 3 min. (MOV 3754 kb)
SUPPLEMENTARY MOVIE 3 Supplementary Movie file 3 shows a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 0.25 mM IPTG at 37 C. The brightfield image is shown
in grey, and fluorescence is shown in green. Total time of movie is 222 min with a sampling rate of one image every 3 min. (MOV 2176 kb) SUPPLEMENTARY MOVIE 4 Supplementary Movie file 4
shows a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 0.5 mM IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green.
Total time of movie is 210 min with a sampling rate of one image every 3 min. (MOV 1033 kb) SUPPLEMENTARY MOVIE 5 Supplementary Movie file 5 shows a timelapse microscopy of JS011 cells
continuously induced with 0.7% arabinose and 0.75 mM IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green. Total time of movie is 268 min with a sampling
rate of one image every 2 min. (MOV 3717 kb) SUPPLEMENTARY MOVIE 6 Supplementary Movie file 6 shows a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 1 mM
IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green. Total time of movie is 176 min with a sampling rate of one image every 2 min. (MOV 2029 kb)
SUPPLEMENTARY MOVIE 7 Supplementary Movie file 7 shows a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 5 mM IPTG at 37 C. The brightfield image is shown in
grey, and fluorescence is shown in green. Total time of movie is 246 min with a sampling rate of one image every 3 min. (MOV 1647 kb) SUPPLEMENTARY MOVIE 8 Supplementary Movie file 8 shows
a timelapse microscopy of JS011 cells continuously induced with 0.7% arabinose and 10 mM IPTG at 37 C. The brightfield image is shown in grey, and fluorescence is shown in green. Total time
of movie is 204 min with a sampling rate of one image every 3 min. (MOV 1538 kb) SUPPLEMENTARY MOVIE 9 Supplementary Movie file 9 shows a timelapse microscopy of JS011 cells continuously
induced with 0.7% arabinose and 2 mM IPTG at 25 C. The phase contrast image is shown in grey, and fluorescence is shown in green. Total time of movie is 702 min with a sampling rate of one
image every 3 min. (MOV 3568 kb) SUPPLEMENTARY MOVIE 10 Supplementary Movie file 10 shows a timelapse microscopy of JS011 cells upon initiation of induction with0.7% arabinose and 2 mM IPTG
at 37 C. The brightfield image is shown in grey, and fluorescenceis shown in green. Total time of movie is 75 min with a sampling rate of one image every 3 min. Note the initial synchrony of
the fluorescence response. (MOV 1078 kb) SUPPLEMENTARY MOVIE 11 Supplementary Movie file 11shows a timelapse microscopy of JS013 cells continuously induced with 0.6mM IPTG at 37 C. The
phase-contrast image is shown in grey, and fluorescence is shown in green. Total time of movie is 210 min with a sampling rate of one image every 3 min. (MOV 2084 kb) SUPPLEMENTARY MOVIE 12
Supplementary Movie file 12 shows a timelapse microscopy of MG1655Z1/pZE12-yemGFP-ssrA cells continuously induced with 2 mM IPTG at 37 C. These cells express GFP from the pLlacO-1 promoter
and express LacI constitutively. There is no feedback control of GFP expression in this strain. The phase-contrast image is shown in grey, and fluorescence is shown in green. Total time of
movie is 252 min with a sampling rate of one image every 3 min. (MOV 2966 kb) POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT
SLIDE FOR FIG. 4 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Stricker, J., Cookson, S., Bennett, M. _et al._ A fast, robust and tunable synthetic
gene oscillator. _Nature_ 456, 516–519 (2008). https://doi.org/10.1038/nature07389 Download citation * Received: 09 July 2008 * Accepted: 05 September 2008 * Published: 29 October 2008 *
Issue Date: 27 November 2008 * DOI: https://doi.org/10.1038/nature07389 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative