Design of multivalent complexes using the barnase·barstar module
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

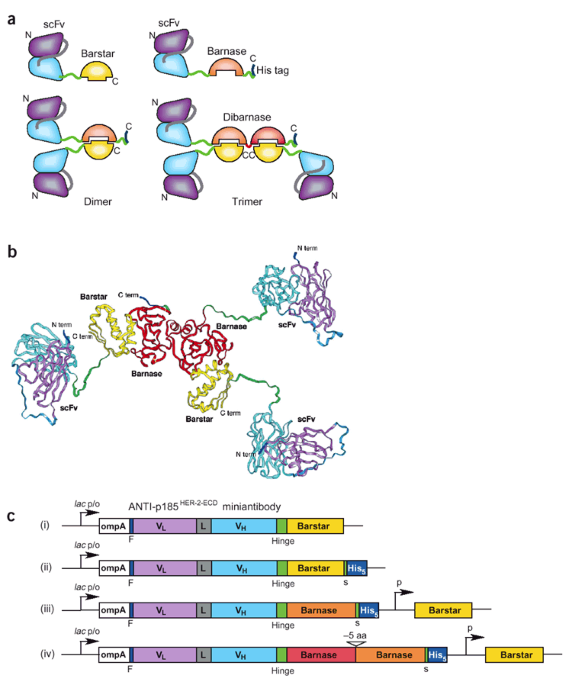
ABSTRACT The ribonuclease barnase (12 kDa) and its inhibitor barstar (10 kDa) form a very tight complex in which all N and C termini are accessible for fusion. Here we exploit this system to
create modular targeting molecules based on antibody scFv fragment fusions to barnase, to two barnase molecules in series and to barstar. We describe the construction, production and
purification of defined dimeric and trimeric complexes. Immobilized barnase fusions are used to capture barstar fusions from crude extracts to yield homogeneous, heterodimeric fusion
proteins. These proteins are stable, soluble and resistant to proteolysis. Using fusions with anti-p185HER2-ECD 4D5 scFv, we show that the anticipated gain in avidity from monomer to dimer
to trimer is obtained and that favorable tumor targeting properties are achieved. Many permutations of engineered multispecific fusion proteins become accessible with this technology of
quasi-covalent heterodimers. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access
to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read
our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SPYMASK ENABLES COMBINATORIAL ASSEMBLY OF BISPECIFIC BINDERS Article Open access 16 March 2024 SELECTION OF
ANTIBODY-BINDING COVALENT APTAMERS Article Open access 08 August 2024 DEVELOPMENT OF MIRROR-IMAGE MONOBODIES TARGETING THE ONCOGENIC BCR::ABL1 KINASE Article Open access 23 December 2024
ACCESSION CODES ACCESSIONS PROTEIN DATA BANK * 1BGS * 1FVC REFERENCES * Batra, S.K., Jain, M., Wittel, U.A., Chauhan, S.C. & Colcher, D. Pharmacokinetics and biodistribution of
genetically engineered antibodies. _Curr. Opin. Biotechnol._ 13, 603–608 (2002). Article CAS Google Scholar * Plückthun, A. & Pack, P. New protein engineering approaches to
multivalent and bispecific antibody fragments. _Immunotechnol._ 3, 83–105 (1997). Article Google Scholar * Todorovska, A. et al. Design and application of diabodies, triabodies and
tetrabodies for cancer targeting. _J. Immunol. Methods_ 248, 47–66 (2001). Article CAS Google Scholar * Bennett, M.J., Schlunegger, M.P. & Eisenberg, D. 3D domain swapping: a
mechanism for oligomer assembly. _Prot. Sci._ 4, 2455–2468 (1995). Article CAS Google Scholar * Dreier, T. et al. Extremely potent, rapid and costimulation-independent cytotoxic T-cell
response against lymphoma cells catalyzed by a single-chain bispecific antibody. _Int. J. Cancer_ 100, 690–697 (2002). Article CAS Google Scholar * Rodrigues, M.L. et al. Engineering
Fab' fragments for efficient F(ab)2 formation in _Escherichia coli_ and for improved _in vivo_ stability. _J. Immunol._ 151, 6954–6961 (1993). CAS PubMed Google Scholar * King, D.J.
et al. Improved tumor targeting with chemically cross-linked recombinant antibody fragments. _Cancer Res._ 54, 6176–6185 (1994). CAS PubMed Google Scholar * Hill, C.P., Anderson, D.H.,
Wesson, L., DeGrado, W.F. & Eisenberg, D. Crystal structure of alpha 1: implications for protein design. _Science_ 249, 543–546 (1990). Article CAS Google Scholar * O'Shea, E.K.,
Klemm, J.D., Kim, P.S. & Alber, T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. _Science_ 254, 543–544 (1991). Article Google Scholar * Jeffrey,
P.D., Gorina, S. & Pavletich, N.P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. _Science_ 267, 1498–1502 (1995). Article CAS Google
Scholar * Pack, P. & Plückthun, A. Miniantibodies: use of amphipathic helices to produce functional, flexibly linked dimeric Fv fragments with high avidity in _Escherichia coli_.
_Biochemistry_ 31, 1579–1584 (1992). Article CAS Google Scholar * de Kruif, J. & Logtenberg, T. Leucine zipper dimerized bivalent and bispecific scFv antibodies from a semi-synthetic
antibody phage display library. _J. Biol. Chem._ 271, 7630–7634 (1996). Article CAS Google Scholar * Terskikh, A.V. et al. “Peptabody”: a new type of high avidity binding protein. _Proc.
Natl. Acad. Sci. USA_ 94, 1663–1668 (1997). Article CAS Google Scholar * Yazaki, P.J. & Wu, A.M. Construction and characterization of minibodies for imaging and therapy of colorectal
carcinomas. _Meth. Mol. Biol._ 207, 351–364 (2003). CAS Google Scholar * Carter, P. Bispecific human IgG by design. _J. Immunol. Methods_ 248, 7–15 (2001). Article CAS Google Scholar *
Hartley, R.W. Barnase-barstar interaction. _Methods Enzymol._ 341, 599–611 (2001). Article CAS Google Scholar * Schreiber, G. Methods for studying the interaction of barnase with its
inhibitor barstar. _Methods Mol. Biol._ 160, 213–226 (2001). CAS PubMed Google Scholar * Schreiber, G. & Fersht, A.R. Rapid, electrostatically assisted association of proteins. _Nat.
Struct. Biol._ 3, 427–431 (1996). Article CAS Google Scholar * Green, N.M. Avidin and streptavidin. _Methods Enzymol._ 184, 51–67 (1990). Article CAS Google Scholar * Guillet, V.,
Lapthorn, A., Hartley, R.W. & Mauguen, Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. _Structure_ 1, 165–177 (1993). Article CAS Google
Scholar * Buckle, A.M., Schreiber, G. & Fersht, A.R. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. _Biochemistry_ 33,
8878–89 (1994). Article CAS Google Scholar * Eigenbrot, C., Randal, M., Presta, L., Carter, P. & Kossiakoff, A.A. X-ray crystal structures of the antigen-binding domains from three
variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. _J. Mol. Biol._ 229, 969–995 (1993). Article CAS Google Scholar * Willuda, J. et al. Tumor
targeting of mono-, di-, and tetravalent anti-p185(HER-2) miniantibodies multimerized by self-associating peptides. _J. Biol. Chem._ 276, 14385–14392 (2001). Article CAS Google Scholar *
Slamon, D. et al. Studies of the HER-2/_neu_ proto-oncogene in human breast and ovarian cancer. _Science_ 244, 707–712 (1989). Article CAS Google Scholar * Yarden, Y. & Sliwkowski,
M.X. Untangling the ErbB signalling network. _Nat. Rev. Mol. Cell Biol._ 2, 127–137 (2001). Article CAS Google Scholar * Waibel, R. et al. Stable one-step technetium-99m labeling of
His-tagged recombinant proteins with a novel Tc(I)-carbonyl complex. _Nat. Biotechnol._ 17, 897–901 (1999). Article CAS Google Scholar * Deyev, S.M., Yazynin, S.A., Kuznetsov, D.A.,
Jukovich, M. & Hartley, R.W. Ribonuclease-charged vector for facile direct cloning with positive selection. _Mol. Gen. Genet._ 259, 379–382 (1998). Article CAS Google Scholar *
Hartley, R.W. Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. _J. Mol. Biol._ 202, 913–915 (1988). Article CAS Google Scholar *
Hartley, R.W. Barnase and barstar: two small proteins to fold and fit together. _Trends Biochem. Sci._ 14, 450–454 (1989). Article CAS Google Scholar * Wörn, A. & Plückthun, A.
Stability engineering of antibody single-chain Fv fragments. _J. Mol. Biol._ 305, 989–1010 (2001). Article Google Scholar * Wörn, A. & Plückthun, A. An intrinsically stable antibody
scFv fragment can tolerate the loss of both disulfide bonds and fold correctly. _FEBS Lett._ 427, 357–361 (1998). Article Google Scholar * Lindner, P. et al. Specific detection of
his-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. _Biotechniques_ 22, 140–149 (1997). Article CAS Google Scholar * Yazaki, P.J. et al. Tumor
targeting of radiometal labeled anti-CEA recombinant T84.66 diabody and T84.66 minibody: Comparison to radioiodinated fragments. _Bioconjugate Chem._ 12, 220–228 (2001). Article CAS Google
Scholar * Nielsen, U.B., Adams, G.P., Weiner, L.M. & Marks, J.D. Targeting of bivalent anti-ErbB2 antibody fragments to tumor cells is independent of the intrinsic antibody affinity.
_Cancer Res._ 60, 6434–6440 (2000). CAS Google Scholar * Casey, J.L. et al. Dosimetric evaluation and radioimmunotherapy of anti-tumour multivalent Fab' fragments. _Br. J. Cancer_ 81,
972–980 (1999). Article CAS Google Scholar * Tahtis, K. et al. Biodistribution properties of (111)indium-labeled C-functionalized trans-cyclohexyl diethylenetriaminepentaacetic acid
humanized 3S193 diabody and F(ab′)(2) constructs in a breast carcinoma xenograft model. _Clin. Cancer Res._ 7, 1061–1072 (2001). CAS PubMed Google Scholar * Trejtnar, F. & Laznicek,
M. Analysis of renal handling of radiopharmaceuticals. _Q. J. Nucl. Med._ 46, 181–194 (2002). CAS PubMed Google Scholar * Ge, L., Knappik, A., Pack, P., Freund, C. & Plückthun, A.
Expressing antibodies in _Escherichia coli_. in _Antibody Engineering_, edn.2 (ed. Borrebaeck, C.A.K.) 229–266 (Oxford University Press, Oxford, 1995). Google Scholar * Knappik, A. &
Plückthun, A. Engineered turns of a recombinant antibody improve its _in vivo_ folding. _Protein Eng._ 8, 81–89 (1995). Article CAS Google Scholar * Hartley, R.W. Directed mutagenesis and
barnase-barstar recognition. _Biochemistry_ 32, 5978–5984 (1993). Article CAS Google Scholar * Bass, S., Gu, Q. & Christen, A. Multicopy suppressors of prc mutant _Escherichia coli_
include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. _J. Bacteriol._ 178, 1154–1161 (1996). Article CAS Google Scholar * Miller, K. et al. Design, construction,
and _in vitro_ analyses of multivalent antibodies. _J. Immunol._ 170, 4854–4861 (2003). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS The authors thank Jörg Willuda for
discussions during the initial phase of this project, Annemarie Honegger for molecular modeling, Stephen F. Marino for help and comments, and Frank Bootz and Lydie Chané-Favre for help in
the immunogenicity experiments. The work was supported by grants from, among others, the Swiss National Science Foundation (no. 7UPJ062274), the Russian Foundation of Basic Research (no.
01-04-49450) and the Russian Science Support Foundation (no. 2077.2003.4) and PCB RAS. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Shemyakin & Ovchinnikov Institute of Bioorganic
Chemistry and Institute of Gene Biology, Russian Academy of Sciences, Miklukho-Maklaya str.16/10, Moscow, 117997, Russia Sergey M Deyev & Ekaterina N Lebedenko * Center of
Radiopharmaceutical Science, Villigen, CH-5232, PSI, Switzerland Robert Waibel & August P Schubiger * Department of Biochemistry, University of Zürich, Winterthurer str. 190, Zürich,
CH-8057, Switzerland Andreas Plückthun Authors * Sergey M Deyev View author publications You can also search for this author inPubMed Google Scholar * Robert Waibel View author publications
You can also search for this author inPubMed Google Scholar * Ekaterina N Lebedenko View author publications You can also search for this author inPubMed Google Scholar * August P Schubiger
View author publications You can also search for this author inPubMed Google Scholar * Andreas Plückthun View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHORS Correspondence to Sergey M Deyev or Andreas Plückthun. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY FIG. 1 (PDF 137 KB) SUPPLEMENTARY METHODS (PDF 162 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Deyev, S., Waibel, R.,
Lebedenko, E. _et al._ Design of multivalent complexes using the barnase·barstar module. _Nat Biotechnol_ 21, 1486–1492 (2003). https://doi.org/10.1038/nbt916 Download citation * Received:
25 August 2003 * Accepted: 25 September 2003 * Published: 23 November 2003 * Issue Date: 01 December 2003 * DOI: https://doi.org/10.1038/nbt916 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative