HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

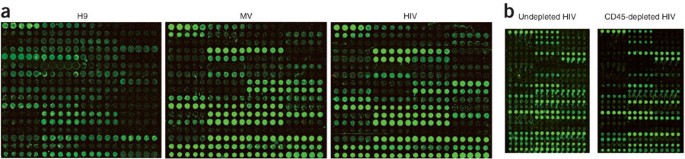
HIV-1 is a master at deceiving the immune system and usurping host biosynthetic machinery. Although HIV-1 is coated with host-derived glycoproteins, only glycosylation of viral gp120 has
been described. Here we use lectin microarray technology to analyze the glycome of intact HIV-1 virions. We show that the glycan coat of human T cell line–derived HIV-1 matches that of
native immunomodulatory microvesicles. The carbohydrate composition of both virus and microvesicles is cell-line dependent, which suggests a mechanism to rapidly camouflage the virus within
the host. In addition, binding of both virus and microvesicles to antiviral lectins is enriched over the host cell, raising concern about targeting these glycans for therapeutics. This work
also sheds light on the binding of HIV-1 to galectin-1, an important human immune lectin. Overall, our work strongly supports the theory that HIV-1 co-opts the exocytic pathway of
microvesicles, thus potentially explaining why eliciting a protective antiviral immune response is difficult.
We thank B. Bohn, J. Miller and B. Imming (AIDS Vaccine Program, NCI-Frederick) for help with virus and microvesicle purification; E. Chertova and D. Roser (AIDS Vaccine Program,
NCI-Frederick) for biochemical analysis of the samples; D. Graham (University of Texas, Austin) for generous use of his ultracentrifuge; L. Baum (UCLA Medical School) for the generous gift
of galectin-1; B. O'Keefe (NCI-Frederick) for the generous gift of cyanovirin, scytovirin and griffithsin; E. Thoyakulathu for help in the lectin analysis; University of Texas at Austin
Microarray Core Facility; and J. Lifson (NCI-Frederick) for insightful reading of the manuscript. We thank Q. Sattentau (University of Oxford) for the Jurkat-Tat-CCR5 cells. In addition, we
wish to acknowledge the Consortium for Functional Glycomics (grant number GM62116) for publicly available glycan array data from their database used in this work. Funding was provided by the
Arnold and Mabel Beckman Foundation (L.K.M.) and the US National Science Foundation (CAREER CHE-0644530, L.K.M.), and by federal funds from the National Cancer Institute, National
Institutes of Health, under contract N01-CO-12400 (J.W.B.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services,
nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Department of Chemistry and Biochemistry, Center for Systems and Synthetic Biology and Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, Texas, USA
AIDS Vaccine Program, National Cancer Institute at Frederick, Frederick, Maryland, USA
Image Analysis Lab, SAIC-Frederick, Inc., National Cancer Institute at Frederick, Frederick, Maryland, USA
L.K. designed and performed research, analyzed data and aided in the writing of the paper. J.W.B. designed and performed research. A.B.P. designed and performed research. K.N. performed
research. L.K.M. designed and performed research, analyzed data and wrote the paper.
Supplementary Figures 1–7, Supplementary Tables 1 and 2, and Supplementary Methods (PDF 7419 kb)
Anyone you share the following link with will be able to read this content: