Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

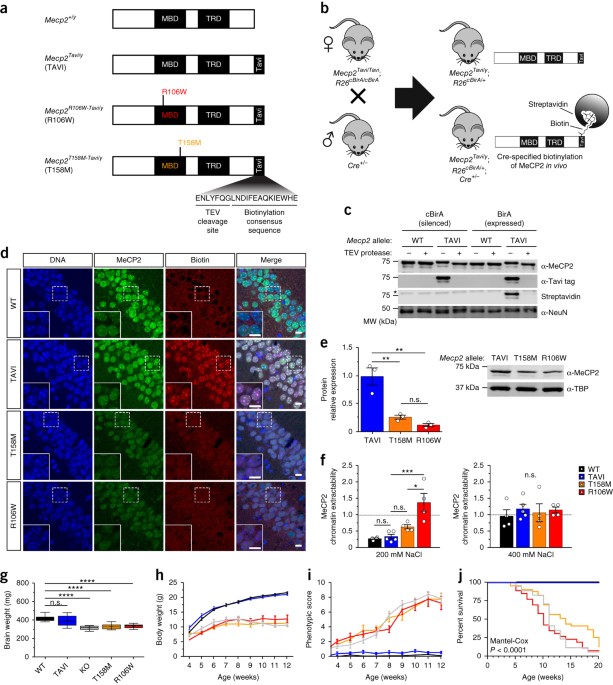
Mutations in MECP2 cause Rett syndrome (RTT), an X-linked neurological disorder characterized by regressive loss of neurodevelopmental milestones and acquired psychomotor deficits. However,
the cellular heterogeneity of the brain impedes an understanding of how MECP2 mutations contribute to RTT. Here we developed a Cre-inducible method for cell-type-specific biotin tagging of
MeCP2 in mice. Combining this approach with an allelic series of knock-in mice carrying frequent RTT-associated mutations (encoding T158M and R106W) enabled the selective profiling of
RTT-associated nuclear transcriptomes in excitatory and inhibitory cortical neurons. We found that most gene-expression changes were largely specific to each RTT-associated mutation and cell
type. Lowly expressed cell-type-enriched genes were preferentially disrupted by MeCP2 mutations, with upregulated and downregulated genes reflecting distinct functional categories.
Subcellular RNA analysis in MeCP2-mutant neurons further revealed reductions in the nascent transcription of long genes and uncovered widespread post-transcriptional compensation at the
cellular level. Finally, we overcame X-linked cellular mosaicism in female RTT models and identified distinct gene-expression changes between neighboring wild-type and mutant neurons,
providing contextual insights into RTT etiology that support personalized therapeutic interventions.
We would like to thank the IDDRC Mouse Gene Manipulation Core at Children's Hospital Boston (U54HD090255, M. Thompson), the Gene Targeting Core (P01DK049210, K. Kaestner) and the Transgenic
and Chimeric Mouse Facility (J. Richa) at University of Pennsylvania for help in generating transgenic mice, the Flow Cytometry and Cell Sorting Resource Laboratory (H. Pletcher, W. DeMuth),
and the Next Generation Sequencing Core (J. Schug) for technical assistance. B.S.J. is supported by a Cell and Molecular Biology Training Grant (TG32GM072290) and the UNCF/Merck Graduate
Research Dissertation Fellowship. This work is supported by NIH grants K22AI112570 (G.V.), R21AI107067 and R01CA140485 (T.H.K.), R01MH091850 and R01NS081054 (Z.Z.), and a basic research
grant from Rettsyndrome.org (Z.Z.). Z.Z. is a Pew Scholar in the Biomedical Sciences.
Brian S Johnson, Ying-Tao Zhao and Maria Fasolino: These authors contributed equally to this work.
Department of Genetics, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA
Brian S Johnson, Ying-Tao Zhao, Maria Fasolino, Janine M Lamonica, George Georgakilas, Kathleen H Wood, Daniel Bu, Yue Cui, Darren Goffin, Golnaz Vahedi & Zhaolan Zhou
Department of Biological Sciences and Center for Systems Biology, University of Texas at Dallas, Richardson, Texas, USA
Conceptualization, B.S.J. and Z.Z.; methodology, B.S.J., J.M.L., D.G. and Z.Z.; investigation, B.S.J., Y.-T.Z., M.F., J.M.L., K.H.W., Y.J.K. and D.B.; formal analyses, B.S.J., Y.-T.Z., G.G.
and T.H.K.; validation, B.S.J., M.F., J.M.L. and G.V.; resources, B.S.J., Y.-T.Z. and Y.C.; data curation, Y.-T.Z.; writing manuscript, B.S.J.; review and editing, B.S.J., Y.-T.Z., M.F. and
Z.Z.; visualization, B.S.J.; project administration and funding acquisition, Z.Z.
Summary of RNA-seq experimental conditions used in this study (XLSX 14 kb)
List of HITS-CLIP data sets used for RBP analysis (XLSX 55 kb)
Anyone you share the following link with will be able to read this content: