Genome engineering using the CRISPR-Cas9 system
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

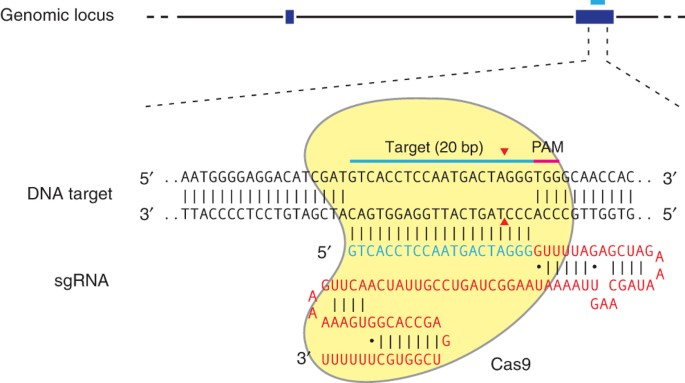
Targeted nucleases are powerful tools for mediating genome alteration with high precision. The RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic
repeats (CRISPR) adaptive immune system can be used to facilitate efficient genome engineering in eukaryotic cells by simply specifying a 20-nt targeting sequence within its guide RNA. Here
we describe a set of tools for Cas9-mediated genome editing via nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in mammalian cells, as well as generation of modified cell
lines for downstream functional studies. To minimize off-target cleavage, we further describe a double-nicking strategy using the Cas9 nickase mutant with paired guide RNAs. This protocol
provides experimentally derived guidelines for the selection of target sites, evaluation of cleavage efficiency and analysis of off-target activity. Beginning with target design, gene
modifications can be achieved within as little as 1–2 weeks, and modified clonal cell lines can be derived within 2–3 weeks.
We thank B. Holmes for help with computational tools. P.D.H. is a James Mills Pierce Fellow and D.A.S. is a National Science Foundation (NSF) pre-doctoral fellow. V.A. is supported by NIH
Training Grants T32GM007753 and T32GM008313. This work was supported by an NIH Director's Pioneer Award (1DP1-MH100706); an NIH Transformative R01 grant (1R01-DK097768); the Keck, McKnight,
Damon Runyon, Searle Scholars, Vallee, Merkin, Klingenstein and Simons Foundations; Bob Metcalfe; and Jane Pauley. Reagents are available to the academic community through Addgene and
associated protocols; support forums and computational tools are available via the Zhang lab website (http://www.genome-engineering.org/).
F Ann Ran and Patrick D Hsu: These authors contributed equally to this work.
Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, Massachusetts, USA
F Ann Ran, Patrick D Hsu, Jason Wright, Vineeta Agarwala, David A Scott & Feng Zhang
McGovern Institute for Brain Research, Cambridge, Massachusetts, USA
Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA.,
Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA
Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts, USA
Program in Biophysics, Harvard University, MIT, Cambridge, Massachusetts, USA
Harvard-MIT Division of Health Sciences and Technology, MIT, Cambridge, Massachusetts, USA
F.A.R., P.D.H., J.W., D.A.S. and F.Z. designed and performed the experiments. V.A. contributed to the online tool. F.A.R., P.D.H. and F.Z. wrote the manuscript with help from all authors.
Anyone you share the following link with will be able to read this content: