Protein lysine crotonylation: past, present, perspective
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Lysine crotonylation has been discovered in histone and non-histone proteins and found to be involved in diverse diseases and biological processes, such as neuropsychiatric disease,
carcinogenesis, spermatogenesis, tissue injury, and inflammation. The unique carbon–carbon π-bond structure indicates that lysine crotonylation may use distinct regulatory mechanisms from
the widely studied other types of lysine acylation. In this review, we discussed the regulation of lysine crotonylation by enzymatic and non-enzymatic mechanisms, the recognition of
substrate proteins, the physiological functions of lysine crotonylation and its cross-talk with other types of modification. The tools and methods for prediction and detection of lysine
crotonylation were also described. SIMILAR CONTENT BEING VIEWED BY OTHERS THE MECHANISMS, REGULATIONS, AND FUNCTIONS OF HISTONE LYSINE CROTONYLATION Article Open access 08 February 2024
ILLUMINATING THE IMPACT OF N-TERMINAL ACETYLATION: FROM PROTEIN TO PHYSIOLOGY Article Open access 15 January 2025 SEQUENCE SPECIFICITY ANALYSIS OF THE SETD2 PROTEIN LYSINE METHYLTRANSFERASE
AND DISCOVERY OF A SETD2 SUPER-SUBSTRATE Article Open access 16 September 2020 INTRODUCTION Protein posttranslational modifications (PTMs) are important epigenetic regulatory mechanisms
involved in diverse biological processes, such as DNA replication, transcription, cell differentiation, and organismal development. Dysregulation of PTMs is associated with a number of
diseases, e.g., neuropsychiatric disease, carcinogenesis, and tissue injury [1]. Due to the development of high-resolution liquid chromatography with tandem mass spectrometry (LC–MS/MS) for
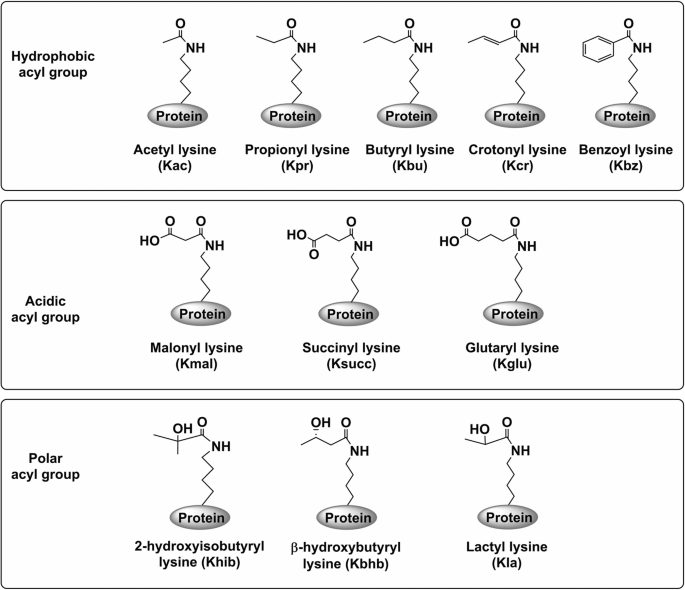
the identification of PTMs, various lysine acylations including acetylation (Kac), butyrylation (Kbu), crotonylation (Kcr), propionylation (Kpr), malonylation (Kmal), glutarylation (Kglu),
benzoylation (Kbz), 2-hydroxyisobutyrylation (Khib), β-hydroxybutyrylation (Kbhb), succinylation (Ksucc), and lactylation (Kla) have been identified [2, 3] (Fig. 1). These modifications
influence protein structure and modulate their stability, localization, and activity [4]. Based on the chemical properties of lysine modification, acylations are classified into three groups
(Fig. 1): the hydrophobic acyl group, the polar acyl group, and the acidic acyl group [1]. Crotonylation was initially identified on lysine residues in histones enriched in the promoter and
enhancer regions in both human somatic and male germinal cells, indicating lysine crotonylation (Kcr) of histone may be an indicator of gene expression [5]. The histone Kcr was conserved
from yeast to human [5]. Subsequently, non-histone crotonylation was identified to be particularly enriched in nuclear proteins involved in RNA processing, nucleic acid metabolism, and
chromosome organization [6]. Later, more studies identified Kcr in non-histone proteins [7,8,9]. The crystal structure of the nucleosome containing crotonylated H3K122cr revealed that
H3K122cr did not affect the overall nucleosome structure, but locally impeded the formation of water-mediated hydrogen bond with DNA backbone, weakened the histone–DNA association, thus
favored the transcriptional activation [10]. Structurally, Kcr is four-carbon in length and the crotonyl modification contains a carbon–carbon (C–C) π-bond that results in a unique rigid
planar conformation [1]. In this review, we will discuss the enzymatic and non-enzymatic regulation of crotonylation, the cellular and physiological functions of Kcr, the cross-talk between
Kcr with other PTMs, and the prediction tools and detection methods for Kcr. REGULATION MECHANISMS OF KCR Protein lysine acylation such as Kcr, Ksucc, Kmal, Kglu, and Kbhb can be regulated
by either enzymatic or non-enzymatic mechanisms [11]. Both serum and urine have been detected with trace amounts of short-chain fatty acid (SCFA) crotonate [12, 13]. Increased crotonate in
colon lumen and serum caused elevated histone Kcr [14]. Supplementation with crotonate dramatically enhanced the levels of cellular crotonyl-CoA and histone Kcr [15]. Besides, treatment with
crotonate significantly increased global Kcr [16], suggesting the abundance of crotonyl-CoA would be one of the main governing factors of Kcr. The process converting crotonate into
crotonyl-CoA was mediated by Acyl-CoA synthetase short chain family member 2 (ACSS2) [15]. Depletion of ACSS2 resulted in drop of cellular crotonyl-CoA and histone Kcr, indicating crotonate
might be the endogenous source of crotonyl-CoA [15]. Besides, the SCFA butyrate through β-oxidation pathway was converted into glutaryl-CoA, and further into crotonyl-CoA by butyryl-CoA
dehydrogenase (BCDH) [17]. Furthermore, the enzymes that catalyze conversion of butyryl-CoA to crotonyl-CoA during fatty acid oxidation, acyl-CoA dehydrogenase short chain (ACADS), and
acyl-CoA oxidase (ACOX3) were key crotonyl-CoA producers during endoderm differentiation [18]. Deletion of ACADS or ACOX3 caused drop of intracellular crotonyl-CoA levels without affecting
other acyl-CoAs [18]. During the amino acid metabolism of lysine, hydroxylysine and tryptophan, glutaryl-CoA dehydrogenase (GCDH) catalyzes the oxidation of glutaryl-CoA to crotonyl-CoA [19,
20]. The GCDH deficiency caused accumulation of glutarylcarnitine and neurotoxic glutaric acid, glutaryl-CoA and 3-hydroxyglutaric acid [21]. Furthermore, chromodomain Y-like (CDYL) was
reported as a crotonyl-CoA hydratase that converts crotonyl-CoA into β-hydroxybutyryl-CoA and negatively regulates histone Kcr [22]. Therefore, these studies support the notion that
crotonyl-CoA, crotonate, and butyrate may drive the occurrence of Kcr (Fig. 2). Besides the regulation of Kcr by intracellular crotonyl-CoA levels, several recent studies have demonstrated
enzyme-regulation on Kcr. The regulation of Kcr is a dynamic balance between the enzymatic activities of writer and eraser proteins that add and remove modification, respectively. The
identification and characterization of writers and erasers is essential for classifying the regulatory mechanisms of protein crotonylation (Table 1, Fig. 3). KCR WRITERS Enzymes that
catalyze modification are referred to as writers. However, crotonyl-specific writers have not been identified yet. Previously characterized histone acetyltransferases (HATs) were shown to
have expanded histone crotonyltransferase (HCT) activities. Three major HAT families including p300/CREB-binding protein (p300/CBP), MYST, and GNAT (Gcn5-related N-acetyltrasferase) were
characterized by their sequences and structures (Supplementary Fig. 1), and have been reported as HCTs that use crotonyl-CoA as substrate to catalyze Kcr [1]. The p300/CBP have both HAT and
HCT activities, and p300-catalyzed histone Kcr can directly stimulate transcription [15]. A hydrophobic pocket, predicted to accommodate the aliphatic portion of remodeled acyl-CoA in the
active site of p300, was observed in the crystal structures of p300 in complex with propionyl-CoA, crotonyl-CoA, or butyryl-CoA [23]. The size of the pocket and its aliphatic nature restrict
against long-chain acyl-CoA variants and instead accommodate short-chain Acyl-CoA such as acetyl-CoA, propionyl-CoA, crotonyl-CoA, or butyryl-CoA without major structural rearrangements
[23]. However, due to the restricted size of an aliphatic back pocket and a substrate-assisted rearrangement of the acyl-CoA chain, the acyltransferase activity of p300 gets weaker with
increasing acyl-chain length [23]. Still, p300/CBP was considered to be the major HCT in mammalian cells [24], the p300/CBP mutants with deficient HAT but competent HCT activity substitute
the endogenous CBP/p300 to enhance transcriptional activation [24]. Later, the global Kcr substrates regulated by p300 were involved in diverse cellular processes [25]. The MYST family
proteins, human MOF, and its yeast homolog Esa1 were detected with a robust HCT activity on both histone H3 and H4 [24]. Deficiency of GNAT family proteins, Gcn5 and HAT1, caused
considerably reduced H3K9cr levels in yeast [26]. However, neither human MOF, yeast Esa1 or yeast Gcn5 displayed any HCT activity in vitro [15, 24, 26], suggesting they may play HCT
activities by forming complex [27, 28]. Indeed, Gcn5 with Ada2 and Ada3 formed ADA complex as a HCT for histone H3Kcr in yeast [29]. Besides, Esa1–Yng2–Epl1 complex was uncovered to function
as histone H3 crotonyltransferase in yeast [29]. Recently, non-histone protein NPM1 was strongly crotonylated by CBP and MOF, and moderately crotonylated by p300/CBP-associated-factor
(PCAF) [9]. However, crotonylation of non-histone protein DDX5 can only be catalyzed by CBP [9]. Non-histone proteins may have distinct HCTs because of their diverse locations. KCR ERASERS
Enzymes that remove modification from specific residues in proteins are referred to as erasers. There are four groups of histone deacetylases (HDACs) [30] (Supplementary Fig. 2). Both class
I and III HDACs were reported as histone decrotonaylases (HDCRs) [2]. HDAC3–NCoR1 complex was first reported to exhibit HDCR activity in vitro [31]. Treatment with histone deacetylase
inhibitors vorinostat and apicidin inhibited the HDCR activity of HDAC3–NCoR1 [31]. Recently, class I HDACs were demonstrated as the major HDCRs in mammalian cells and displayed distinct
site specificity from histone decrotonylation by class III HDAC (SIRT1) [16]. Given that class I HDACs exhibited a major HDCR activity while class II HDACs were deficient in HDCR activity,
key residues of catalytic centers in class I and II HDACs were aligned and major differences were identified [16]. HDAC1 and HDAC3 mutants that lose HDAC but keep intact HDCR activity
displayed a global transcriptional repression and diminished the promoter association with crotonylated histones [16]. Recently, HDAC1-3 regulated HDCR in colon in response to SCFA generated
by microbiota of the gut [14]. Genetic deletion of HDAC1/2 in embryonic stem cells (ESCs) increased global histone crotonylation and resulted in 85% reduction in total HDCR activity [32].
The Class III HDACs, SIRT1-2 were acting as efficient HDACs [33]. By an optimized cross-linking assisted and stable isotope labeling of amino acids in cell culture-based protein
identification approach to comprehensively profile erasers that recognize histone Kcr marks, human SIRT1-3 were identified as HDCRs [34]. The crystal structure of human SIRT3–H3K4cr complex
was solved and the crotonyl-lysine of H3K4cr was located in a hydrophobic pocket of SIRT3 [34]. Residue His248 interacted with the crotonyl amide oxygen via hydrogen bonding and the phenyl
ring of residue Phe180 aligned parallel to the planar crotonyl group and formed π–π stacking interaction with the C–C double bond of crotonyl-lysine [34]. Alignment of all sirtuins
demonstrated that the residue Phe180 of SIRT3 is conserved in SIRT1-2, but not in other sirtuins, which may explain why SIRT4-7 were not identified as HDCRs [34]. However, the levels of
histone crotonylation were higher in SIRT3 lacking cells, but not in those lacking SIRT1/2, suggesting endogenous SIRT3 as a main HDCRs [34]. Recently, crotonylated NPM1 was increased after
a pan-HDAC inhibitor TSA treatment, suggesting HDACs may influence NPM1 crotonylation [9]. HDAC1 and HDAC3, but not HDAC2, decrotonylated NPM1, which can be reversed upon TSA treatment [9].
KCR READERS The level of Kcr could be influenced by the levels of intracellular crotonyl-CoA, and the ratio of crotonyl-CoA/acetyl-CoA, as well as the dynamic balance between
crotonyltransferase and decrotonylase [2]. Thus, the function of Kcr modification in physiology and pathology may be dependent on the readers that recognize Kcr modification. For the
well-studied histone Kac, three major families of readers have been characterized: bromodomain proteins, YEATS domain proteins, and double plant homeodomain finger (DPF) proteins [35]
(Supplementary Fig. 3). Although a subset of bromodomain-containing proteins such as BRD9 and TAF1 were shown to recognize Kcr, their binding affinities are much weaker with crotonylated
peptides than with acetylated peptides [36]. On contrary, DPF or YEATS domain proteins displayed preference for histone Kcr to other types of acylation [37, 38]. Recent studies demonstrated
that the YEATS domain more favors Kcr than Kac [26, 37, 39]. Calorimetric titrations revealed that AF9 YEATS possesses a 2.4-fold binding enhancement for Kcr over Kac, and this favorable Kcr
readout was conserved in human ENL, yeast Yaf9 and Taf14 [37]. The crystal structure of AF9 YEATS in complex with H3K9cr revealed that AF9 YEATS use the same Kac-binding aromatic sandwich
cage for Kcr recognition, with only slight conformational changes of aromatic residues [37]. Besides the hydrogen bonding interactions, preferential binding to Kcr is notably contributed by
π-aromatic interactions of the planar crotonylamide group with aromatic rings in the AF9 binding pocket [37]. By comparison between the crystal structures of BRD3-H3K18ac and AF9-H3K18cr,
the mechanism of YEATS as preferential Kcr reader was displayed [36]. Kcr is too rigid for the reader pockets of most BRD proteins except for those that have a wider pocket, such as TAF1
[36]. However, the elongated and end-open reader packet of YEATS is ideal for interaction with acyl chains of Kcr. This unique mechanism [40] was also observed in human YEATS2 [39] and yeast
Taf14 [26]. By targeting the π–π–π stacking in the aromatic ‘sandwich’ cage, a set of YEATS inhibitors were developed [41,42,43,44,45,46]. DPF domain proteins, including MOZ, MORF, and
DPF1-3, were characterized as Kac readers [1]. Recently, the DPF domain was characterized as histone Kcr-preferential reader [38]. DPF domains of MOZ and DPF2 displayed 4 to 8-fold binding
enhancement of Kcr over Kac [38]. The crystal structure of DPF domain of MOZ in complex with H3K14cr peptide revealed that a hydrophobic ‘dead-end’ pocket lacking aromatic sandwiching
residues accommodated Kcr [38]. Notably, hydrophobic ‘dead-end’ pocket with selectivity for crotonylation was originated from intimate encapsulation and an amide-sensing hydrogen bonding
network [38]. Therefore, the histone Kcr was recognized by π–π–π stacking mechanism of the YEATS domain and intimate hydrophobic ‘dead-end’ mechanism of the DPF domain. THE FUNCTIONS OF KCR
IN PHYSIOLOGY AND PATHOLOGY Several recent studies have demonstrated that Kcr is implicated in various physiological processes [5, 15] (Fig. 3). DNA DAMAGE AND REPAIR The level of H3K9cr
exhibited rapid and transient decrease at DNA damage sites following DNA damage by exposing to laser-microirradiation, ionizing radiation, ultraviolet radiation or by treatment with
etoposide damaging agents [47]. HDACs, but not SIRTs mediated the reduction in H3K9cr during DNA damage [47]. On the other hand, the level of RPA1 Kcr was upregulated upon DNA-damaging and
was negatively regulated by CDYL1 [22]. The Kcr modification of RPA1 enhanced the interaction of RPA1 with single-stranded DNA and with components of resection machinery, and facilitated
cell survival under DNA damage conditions [22]. Although the study indicated that CDYL reduced Kcr of RPA1, the possibility that RPA1 Kcr could be regulated by other factors such as HCTs
and/or HDCRs could not be ruled out. These unidentified factors and CDYL together may contribute to the dynamics of RPA1 Kcr upon DNA-damaging. NEUROPSYCHIATRIC DISEASE Under chronic social
defeat stress and micro-defeat stress, lower level of histone Kcr was exhibited in the medial prefrontal cortex concurrent with selective upregulation of CDYL [48]. Furthermore, _Cdyl_
expression in prelimbic cortex influenced the stress-induced depression-like behaviors in mice [48]. Subsequently, CDYL regulated stress-induced depression-like behaviors by inhibiting VGF
nerve growth factor-mediated transcription, and this activity of CDYL was dependent on its dual hydratase function on histone Kcr and H3K27me3 at the VGF promoter [48]. Thus, CDYL-mediated
reduction of histone Kcr played a critical role in regulating stress-induced depression [48]. Although lack of site-specific histone Kcr antibodies and mutants made it unable to specifically
interrogate the function of Kcr, the observation that histone Kcr may affect major depressive disorders uncovered a possible regulatory mechanism that contributes to this neuropsychiatric
disease. In Alzheimer’s disease (AD), nuclear paraspeckle assembly transcript 1 (NEAT1), a long non-coding RNA, mediated the autoacetylation of p300, which altered the level of H3K27ac and
H3K27cr and the transcription of endocytosis-related genes [49]. The low level of acetyl-CoA after NEAT1 inhibition caused decrease of H3K27ac and increase of H3K27cr [49]. This distinct
alteration reveals the different roles of H3K27ac and H3K27cr in regulation of gene expression, which provides insight on the epigenetic regulatory mechanism of NEAT1 in AD pathology [49].
SELF-RENEWAL AND DIFFERENTIATION OF STEM CELLS Histone Kcr was detected with much higher levels in mouse ESCs [16]. Induced HDAC1-VRPP mutant with intact HDCR but impaired HDAC activity
caused marked reduction of histone Kcr and a drastic reduction of the ESC pluripotency factors, and an increase of endoderm, mesoderm, and ectoderm markers [16]. Thus, enriched histone Kcr
was required for self-renewal of ESCs [16]. Recently, an enrichment of both H3K18ac and H3K18cr at bivalent genes upon deletion of HDAC1-2 in ESCs was observed [32], suggesting a role of
HDAC1-2 in controlling the developmentally regulated genes prior to ES cell differentiation. Consistently, top 10% of genes enriched for either H3K18cr or H3K18ac upon HDAC1-2 deletion were
functional in embryonic morphogenesis and embryo development [32]. Sufficient telomere lengths contribute to unlimited self-renewal and genomic stability of pluripotent stem cells (PSCs)
[50, 51]. Crotonic acid-induced histone Kcr may protect telomeres by activating two-cell genes and Zscan4 and increasing T-SCE-based ALT-like activity [52]. Moreover, Kcr enhances the
efficiency of chemical induction of pluripotent cells [52], although more experiments are needed to understand whether crotonylation directly or indirectly regulates the induction process.
Recently, during differentiation of ESCs, key crotonyl-CoA-producing enzymes such as ACSS2, ACADS, and ACOX3 were significantly induced and enriched in endoderm and/or mesoderm
differentiation, indicating endoderm differentiation is associated with increased crotonyl-CoA production [18]. Histone crotonylation and endodermal gene expression were enhanced upon
differentiation of endoderm [18]. Furthermore, endoderm differentiation was promoted by crotonate, and disrupted histone crotonylation by deletion of crotonyl-CoA-producing enzymes impaired
meso/endoderm differentiation [18]. Most recently, systematic crotonylome profiling in mouse PSCs in different states displayed that majority of crotonylated proteins were involved in
pluripotency-related pathways such as RNA biogenesis, central carbon metabolism, and proteasomal degradation [53]. High crotonyl-CoA levels by adding crotonic acid promoted proteasome
activities in metastable PSCs and facilitated sustaining of pluripotency [53]. HIV LATENCY Epigenetic regulation of histone tails at the human immunodeficiency virus (HIV) long-terminal
repeat is essential for the establishment, maintenance, and reactivation of HIV latency [54]. Elevated histone Kcr by ACSS2 at the HIV LTR caused the reactivation of latent HIV and viral
transcription [55], suggesting its potential role in HIV latency establishment. Besides, a remarkable synergistic reactivation of latent HIV arises when ACSS2-induced histone Kcr is combined
with either PKC agonist PEP005, or vorinostat [55]. Besides, high level of ACSS2 in intestinal mucosa was correlated with altered fatty acid metabolism in the simian immunodeficiency
virus-infected non-human primate models of AIDS [55]. CARCINOGENESIS Histone H3K18cr was the most abundant histone Kcr in intestine, especially in the TSS of colon epithelial crypts [14].
SCFAs are the main products of gut microbiota and affect cellular metabolism and gene transcription in intestine. Depletion of the gut microbiota of mice with antibiotics not only led to a
drop in luminal and serum SCFAs, but also caused an increased expression of HDAC2 and decline of histone Kcr in colon [14]. Besides, bioinformatics analysis revealed that high level of
H3K18cr was involved in cancer [14]. Gut microbiota modulated carcinogenesis via various manners [56], and these above studies suggested that dysregulation of gut microbiota may affect
carcinogenesis by altering histone Kcr. Future studies may focus on the regulation mechanism of microbiota, SCFAs, and histone Kcr in modulating carcinogenesis. Later, crotonylome
alterations by p300 were involved in nonsense-mediated decay, infectious disease, and viral/eukaryotic translation pathways [25]. Additionally, 4.5% of the cancer protein biomarkers in the
Early Detection Research Network database were crotonylated [25]. 5.9% of total genes in the Catalogue of Somatic Mutations in Cancer cancer gene database were found to encode proteins
crotonylated by p300 [25]. Six p300-targeted crotonylated proteins were confirmed as cancer-related proteins [25]. Crotonylated proteins were widely expressed in human tumor tissues [57].
The global Kcr was decreased in liver, stomach, and kidney carcinomas, and elevated in thyroid, esophagus, colon, pancreas, and lung carcinomas [57]. This indicated Kcr may play diverse
roles in cancer progression by modulating different pathways. Changes in global Kcr may partially reflect its association with cancer progression; however, more specific and critical
crotonylation factors regulating cancer progression are waiting for unearthing. SPERMATOGENESIS An intense labeling of histone Kcr was observed in post-meiotic male germ cells and was
related with X-linked haploid cell-specific gene expression program, indicating a role of histone Kcr in epigenetic modification in the post-meiotic stages of spermatogenesis [5]. Besides,
the negative regulation of histone Kcr by CDYL contributed to transcriptional repression and affected the reactivation of sex chromosome-linked genes in round spermatids and the genome-wide
histone replacement in elongating spermatids [58]. In _Cdyl_ transgenic mice, the dysregulation of histone Kcr by _Cdyl_ was associated with reduction of male fertility with a decreased
epididymal sperm count and sperm cell motility [58], implicating CDYL-regulated histone Kcr alteration played an essential role in spermatogenesis. Most recently, Kcr was significantly
enriched at H3K27 compared to Kac during mouse spermatogenesis [59]. Besides, a combined high level of H3K27ac and H3K27cr existed in super-enhancers determined in spermatocytes and round
spermatids [59]. TISSUE INJURY Histone Kcr levels were increased in mouse kidney tissue during acute kidney injury (AKI) induced by folic acid or cisplatin treatment [60]. The increased
histone Kcr in mouse kidney tissue during AKI was associated with increased PGC-1a and SIRT3 and decreased CCL2 [60]. Furthermore, after adding crotonate in cultured tubular cells or
intraperitoneal injection of crotonate, high level of Kcr elevated the expression of PGC-1a and SIRT3 and enhanced protection from AKI [60]. Thus, crotonate may have a potential therapeutic
effect on kidney damage, specifically in AKI by increasing histone Kcr [60]. INFLAMMATION By utilizing the LPS-induced inflammatory response in RAW264.7, histone Kcr was enhanced by
supplement with crotonate prior to LPS stimulation [15]. However, knockdown of ACSS2 resulted in decreased histone Kcr and expression of inflammatory genes upon LPS stimulation [15]. The
recruitment of YEATS domain protein AF9 to LPS-induced genes was enhanced by crotonate pre-treatment in a YEATS-dependent manner [37]. Knockout of AF9 significantly reduced the crotonate
response to LPS stimulation but did not abolish it completely, suggesting other Kcr reader(s) may be also involved in this response [37]. CARDIOVASCULAR DISEASES In human cardiac
hypertrophy, short-chain enoyl-CoA hydratase (ECHS1) was reduced, which was coupled with elevated H3K18cr and H2BK12cr. Deficiency of ECHS1 markedly increased H3K18cr, H2BK12cr, and NFATc3
levels, which further drove the expression of hypertrophic fetal genes and finally promoted the hypertrophic growth of neonatal cardiomyocytes, indicating the essential role of ECHS1 and
histone crotonylation in maintaining the maturity and homeostasis of cardiomyocytes [61]. THE FUNCTIONS OF KCR IN PLANTS After initial identification of Kcr [5], crotonylome analysis in
tobacco [7], papaya fruit [62], rice [63], and peanut [64] have been reported (Table 2). In rice, Kcr and Kbu were enriched as histone modification marks that regulate gene expression [65]
(Table 2). Under starvation or submergence, Kcr and Kbu displayed less dynamic compared to H3K9ac, indicating these modifications may display distinct responses to external and internal
signals and may represent novel epigenetic mechanisms to fine-tune gene expression for plant adaptation [65]. In response to low temperature, temperature-induced lipocalin-1-like (DgTIL1)
was crotonylated, which prevented the degradation of nonspecific lipid transfer protein (DgnsLTP). DgnsLTP then promoted expression and activity of POD, which decreased the accumulation of
ROS under cold stress and promoted the cold resistance of chrysanthemum [66]. Besides, crotonylome analysis in chrysanthemum at low temperature identified 393 upregulated and 500
downregulated proteins [67]. Furthermore, crotonylated ascorbate peroxidase (APX) increased APX activity and further reduced the oxidative damage caused by low-temperature stress [67] (Table
2). In addition, various crotonylated proteins in tea plants under NH4+ deficiency/resupply were found to participate in diverse biological processes such as photosynthesis, carbon
fixation, and amino acid metabolism [68] (Table 2), suggesting a profound role of Kcr on the metabolic processes in tea leaves. THE FUNCTIONS OF KCR IN MICROBIOLOGY The conserved histone Kcr
was detected in yeast _Saccharomyces cerevisiae_ [5]. Yeast HATs (Gcn5, Rtt109, and HAT1) and HDACs (Rpd3, Hos1, and Hos2) were identified as crotonyltransferases and decrotonylases for
their function in regulating H3K9cr levels [26]. In addition, yeast Yaf9 and Taf14 were found as Kcr readout [26, 37]. During yeast metabolic cycle (YMC), the periodical expression of fatty
acid β-oxidation genes was coincident with histone crotonylation. During nutrient limitation, H3K9cr peaked while K3K9ac declined, and expression of pro-growth genes was prohibited [69].
Adding of crotonic acid elevated the Kcr levels and the constitutive repression of pro-growth genes and caused the disruption of YMC oscillation [69]. Yeast Taf14 was necessary for the
transcriptional oscillation of YMC [69]. Besides, Yeast Taf14 was participated in PIC stabilization and was required for yeast survival [70]. In _Streptomyces roseosporus_, Kcr upregulated
carbon catabolite repression metabolism by negative regulating the activity of glucose kinase Glk and the utilization of carbon sources [71]. Kcr level and Glk activity were modulated by
decrotonylase CobB and crotonyltransferase Kct1 [71] (Table 2). Histone crotonylation in pathogenic _Candida albicans_ was dynamically controlled by metabolism and stress responses [72].
Crotonate can regulate responsive transcriptional program and result in resistance against cell wall stress [72]. Taf14 is essential for _Candida albicans_ virulence by controlling gene
expression, stress resistance and invasive growth via its chromatin reader function [72]. Crotonylome analysis in _Candida albicans_ displayed that majority of crotonylated proteins were
involved in biosynthetic events and carbon metabolism [73]. After treatment with patulin, 79 upregulated crotonylated proteins were involved in tricarboxylic acid cycle and gluconeogenic
pathway and 46 downregulated crotonylated proteins were related with ribosome and carbohydrate transport and metabolism, which predicted the role of Kcr in patulin degradation [74]. THE
IDENTIFICATION AND DETECTION OF KCR EXPERIMENTAL METHODS Due to the development of high-sensitivity mass spectrometry, new PTMs could be identified. Unbiased, systematic screenings have been
applied to discover new lysine acylations [1]. A pan antibody against Kcr was generated to directly detect Kcr [5]. Isotopic labeling, previously used for the detection of Kac [75], was
also used to detect Kcr [5]. Xie et al. developed the genetically encoded photoaffinity analogues of Kcr that can site-specifically incorporate into proteins via the genetic code expansion
strategy [76]. The crotonyl mark is highly reactive toward phosphine nucleophiles that contain a pendent carboxylic acid group [77]. Based on water-soluble phosphine warhead, a covalent
chemo-proteomic probe for the detection and functional analysis of Kcr was developed, allowing detection of endogenous cellular proteins being crotonylated [77]. Most recently, single-step
fluorescent probes (KTcr-I that is recognized by Sirt2, and KTcr-II that is recognized by HDAC3), which generate fluorescence signal by intramolecular nucleophilic exchange reaction, to
detect decrotonylation activity of HDACs were developed [78]. Although extensive structural and mechanistic studies, the cross regulation between different types of acylations remains
unclear. In order to clarify the role of different lysine acylations, development of acyl-type specific enzymes would be a useful tool. For this, p300 I1935G and CBP I1432G mutants with
deficient HAT but competent HCT activities [24] and HDAC1/3 AGG-VRPP mutant with lacking of HDCR but intact HDAC activities [16] were generated. Most recently, relying on replacing an
essential active-site lysine residue of orotidine-5’-monophosphate decarboxylase with lysine derivatives by genetic code expansion, a selection system for HDAC-HDCR was designed in yeast
[79]. BIOINFORMATICS TOOLS Experimental approaches for identifying Kcr sites are often time-consuming and labor-intensive, thus difficult to widely popularize in large-scale species. On the
other hand, computational approaches are cost-effective and can be used in a high-throughput manner to generate relatively precise identification. A discrete hidden Markov model implemented
with a software named CrotPred for predicting Kcr sites was established [80]. Then, a new approach to predict Kcr sites based on support vector machine was presented [81]. To improve the
performance of the computational prediction of crotonylation sites, CKSAAP CrotSite was developed [82]. Based on the CKSAAP CrotSite model, whose sensitivity reached 92.45%, a user-friendly
web-server was established [82]. In the meanwhile, a user-friendly web-server named iKcr-PseEns by incorporating five tiers of amino acid pairwise couplings into the general pseudo amino
acid composition was also built [83]. Malebary et al. proposed an improved Kcr predictor named iCrotoK-PseAAC, in which various position and composition relative features along with
statistical moments were incorporated in this predictor [84]. Later, based on physicochemical property and evolutionary-derived feature of protein sequences, LightGBM-CroSite was developed
[85]. Most recently, Lv et al. performed a deep learning-based method termed Deep-Kcr by combining sequence-based features, physicochemical property-based features and numerical
space-derived information [86]. Although these tools showed powerful prediction, experimental approaches need to be employed to confirm the prediction results. Since experimental approaches
are visible to reflect the dynamics of modification, efficient, lab-common, and inexpensive experimental approaches are urgently needed. KCR VERSUS KAC The overlap between histone Kcr and
Kac [5] (Fig. 4), raised the possibility of crosstalk between these two PTMs. In Alzheimer’s disease, NEAT1 promoted the autoacetylation of P300 and its acyltransferase activity, and altered
the level of H3K27ac and H3K27cr simultaneously [49]. During YMC, both histone crotonylation and acetylation dynamically fluctuated and this fluctuation had distinct peaks at different
points in the metabolic cycle [69]. Although Kac and Kcr shared modulators such as writers, erasers, and readers, Kcr may use distinct regulatory modulators from Kac due to the presence of
C–C π-bond. Crotonyl group has a more rigid structure. However, the acetyl group is tetrahedral and rotatable. Indeed, YEATS and DPF domain had enhanced binding affinity for Kcr over Kac
[37, 38]. Both Kcr and Kac were critical for global transcription in mammalian cells [16]. However, Kcr was reported to preferentially ‘escapee genes’ during post-meiotic sex inactivation in
mouse testis [5]. In addition, p300-mediated histone Kcr displayed greater stimulation on gene transcription than histone Kac [15]. CBP/p300 mutants with deficient HAT and intact HCT
activity [24] and HDAC1/3 mutants with impaired HDAC but intact HDCR activities [9] indicated different modulation patterns between Kac and Kcr. In distinct metabolic conditions, or
different progression status of tissues, the patterns of Kcr and Kac were different, which may be due to the altered concentrations of distinct CoA [5]. CONCLUSIONS AND PERSPECTIVES Kcr is a
recently identified posttranslational modification that occurs in a wide range of proteins both in prokaryotes and eukaryotes [5,6,7, 26, 65, 71, 72, 87,88,89] (Table 2). Although Kcr has
been shown to be involved in diverse cellular functions in health and disease situations, the underlying mechanism of Kcr in these biological processes are unclear. Aberration in
crotonylation and decrotonylation was associated with several diseases. Thus, one of the future focuses may be the in-depth understanding of the substrates targeted by Kcr, and their
biological roles regulated by this modification. Cellular concentration of crotonyl-CoA influenced histone and non-histone Kcr and further altered biological processes [15]. Reports
demonstrated that ACSS2 and CDYL regulate the level of crotonyl-CoA in tissues and in cells [15, 22]. Therefore, one angle to clarify the function of Kcr in biological processes is to
measure crotonyl-CoA levels in tissues and in subcellular compartments and identify factors that influence the levels of crotonyl-CoA. The number of enzymes that catalyze or hydrolyze Kcr is
still few. In addition, the Kcr writers, erasers, and readers are generally shared with other PTMs and it is unclear whether specific enzymes for Kcr exist. Therefore, identifying these
specific enzymes for Kcr would be interesting. The overlap between Kcr and other PTMs, such as Kcr and Kac [5], aroused consideration on whether different acylations have unique regulatory
roles or they perform redundant functions. Besides, investigation into the relative stoichiometries of various acylations occurring on the same lysine residue would be an interesting aspect
for future studies. FACTS * Lysine crotonylation (Kcr) is newly identified protein posttranslational modification in histone and non-histone proteins, and is involved in diverse diseases and
biological processes, such as neuropsychiatric disease, carcinogenesis, spermatogenesis, tissue injury and inflammation, by influencing protein structure and modulate protein stability,
localization, and activity. * The unique carbon–carbon (C–C) π-bond structure of Kcr resulting in a rigid planar conformation indicates distinct regulatory mechanisms from the widely studied
other types of lysine acylation. * The intensity of Kcr could be influenced by the levels of intracellular crotonyl-CoA, the ratio of crotonyl-CoA/acetyl-CoA, as well as the dynamic balance
between crotonyltransferase and decrotonylase. * The functions of Kcr in physiology and pathology are dependent on the readers that recognize Kcr modification. YEATS and DPF domain proteins
have been characterized as histone Kcr-preferential reader. * The overlap between histone Kcr and Kac raises the possibility of crosstalk between these two PTMs that display distinct roles
in the same disease and biological process. * Experimental and computational approaches have been developed for prediction, identification, and analyzing the regulatory mechanisms of Kcr.
OPEN QUESTIONS * Although Kcr has been shown to be involved in diverse cellular functions in health and disease situations, the underlying mechanism of Kcr roles are unclear. Thus, one of
the future focuses may be the in-depth understanding of the substrates targeted by Kcr, and their biological roles regulated by this modification. * How to measure crotonyl-CoA levels in
tissues and in subcellular compartments? What are factors that influence the levels of crotonyl-CoA in tissues and subcellular compartments? * Whether specific enzymes that catalyze or
hydrolyze histone Kcr exist? Whether non-histone Kcr uses distinct crotonyltransferases and decrotonylases from histone Kcr because of their diverse locations? * Whether readout of
non-histone kcr shares similar recognition mechanisms as histone Kcr? * The overlap between Kcr and other PTMs, such as Kcr and Kac, arouses consideration on whether different acylations
have unique regulatory roles or they perform redundant functions. Besides, investigation into the relative stoichiometries of various acylations occurring on the same lysine residue would be
an interesting aspect for future studies. * The efficient, lab-common, and inexpensive experimental approaches for Kcr detection are urgently needed. For clarifying the roles of different
lysine acylations, development of acyl-type specific enzymes would be helpful. REFERENCES * Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone
acylations. Nat Rev Mol Cell Biol. 2017;2:90–101. Article CAS Google Scholar * Zhao S, Zhang X, Li H. Beyond histone acetylation-writing and erasing histone acylations. Curr Opin Struct
Biol. 2018;53:169–77. Article CAS PubMed Google Scholar * Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature.
2019;574:575–80. * Figlia G, Willnow P, Teleman AA. Metabolites regulate cell signaling and growth via covalent modification of proteins. Developmental Cell. 2020;54:156–70. Article CAS
PubMed Google Scholar * Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification.
Cell. 2011;146:1016–28. Article CAS PubMed PubMed Central Google Scholar * Wei W, Mao A, Tang B, Zeng Q, Gao S, Liu X, et al. Large-scale identification of protein crotonylation
reveals its role in multiple cellular functions. J. Proteome Res. 2017;4:1743–52. Article CAS Google Scholar * Sun H, Liu X, Li F, Li W, Zhang J, Xiao Z, et al. First comprehensive
proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci Rep. 2017;7:3013–26. Article PubMed PubMed Central CAS Google Scholar * Wu Q, Li W, Wang C, Fan P,
Cao L, Wu Z, et al. Ultradeep lysine crotonylome reveals the crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J Proteome Res. 2017;16:3664–71. Article
CAS PubMed Google Scholar * Xu W, Wan J, Zhan J, Li X, He H, Shi Z, et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27:946–9. Article PubMed PubMed
Central Google Scholar * Suzuki Y, Horikoshi N, Kato D, Kurumizaka H. Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122. Biochem Biophys Res Commun.
2016;469:483–9. Article CAS PubMed Google Scholar * Simic Z, Weiwad M, Schierhorn A, Steegborn C, Schutkowski M. The ɛ-amino group of protein lysine residues is highly susceptible to
nonenzymatic acylation by several physiological Acyl-CoA thioesters. Chembiochem. 2015;16:2337–47. Article CAS PubMed Google Scholar * Kang H, Li X, Zhou Q, Quan C, Xue F, Zheng J, et
al. Exploration of candidate biomarkers for human psoriasis based on gas chromatography-mass spectrometry serum metabolomics. Br J Dermatol. 2017;176:713–22. Article CAS PubMed Google
Scholar * Herter CA. The acid intoxication of diabetes in its relation to prognosis. J Exp Med. 1901;6:617–33. Article Google Scholar * Fellows R, Denizot J, Stellato C, Cuomo A, Jain P,
Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. 2018;9:105–19. Article PubMed PubMed
Central CAS Google Scholar * Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, et al. Intracellular Crotonyl-CoA stimulates transcription through p300-catalyzed histone
crotonylation. Mol Cell. 2015;58:203–15. Article CAS PubMed PubMed Central Google Scholar * Wei W, Liu X, Chen J, Gao S, Lu L, Zhang H, et al. Class I histone deacetylases are major
histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. Article CAS PubMed PubMed Central Google Scholar *
Jeong J, Bertsch J, Hess V, Choi S, Choi I, Chang IS, et al. Energy conservation model based on genomic and experimental analyses of a carbon monoxide-utilizing, butyrate-forming acetogen,
_Eubacterium limosum_ KIST612. Appl Environ Microbiol. 2015;81:4782–90. Article CAS PubMed PubMed Central Google Scholar * Fang Y, Xu X, Ding J, Yang L, Doan MT, Karmaus PWF, et al.
Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell. 2021;28:748–763. * Wu L, Qiao Y, Gao J, Deng G, Yu W, Chen G, et al. Functional
characterization of rat glutaryl-CoA dehydrogenase and its comparison with straight-chain acyl-CoA dehydrogenase. Bioorg Medicinal Chem Lett. 2011;21:6667–73. Article CAS Google Scholar *
Dwyer TM, Rao KS, Goodman SI, Frerman FE. Proton abstraction reaction, steady-state kinetics, and oxidation-reduction potential of human Glutaryl-CoA dehydrogenase†. Biochemistry.
2000;39:11488–99. Article CAS PubMed Google Scholar * Biagosch C, Ediga RD, Hensler S, Faerberboeck M, Kuehn R, Wurst W, et al. Elevated glutaric acid levels in Dhtkd1-/Gcdh- double
knockout mice challenge our current understanding of lysine metabolism. Biochim et Biophys Acta. 2017;9:2220–8. Article CAS Google Scholar * Yu H, Bu C, Liu Y, Gong T, Liu X, Liu S, et
al. Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair. Sci Adv. 2020;6:y4697. Article CAS Google Scholar * Kaczmarska Z, Ortega
E, Goudarzi A, Huang H, Kim S, Márquez JA, et al. Structure of p300 in complex with acyl-Coa variants. Nat Chem Biol. 2017;1:21–9. Article CAS Google Scholar * Liu X, Wei W, Liu Y, Yang
X, Wu J, Zhang Y, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and
crotonyltransferase-competent CBP/p300. Cell Discov. 2017;3:17016–32. Article CAS PubMed PubMed Central Google Scholar * Huang H, Wang D, Zhao Y. Quantitative crotonylome analysis
expands the roles of p300 in the regulation of lysine crotonylation pathway. Proteomics. 2018;18:1700230. Article CAS Google Scholar * Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest
A, Tsun IK, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12:396–8. Article CAS PubMed PubMed Central Google Scholar * Decker PV, Yu DY,
Iizuka M, Qiu Q, Smith MM. Catalytic-site mutations in the MYST family histone acetyltransferase Esa1. Genetics. 2008;178:1209–20. Article CAS PubMed PubMed Central Google Scholar *
Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, et al. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci. 1998;95:3561–5. Article CAS
PubMed PubMed Central Google Scholar * Kollenstart L, de Groot AJL, Janssen GMC, Cheng X, Vreeken K, Martino F, et al. Gcn5 and Esa1 function as histone crotonyltransferases to regulate
crotonylation-dependent transcription. J Biol Chem. 2019;294:20122–34. Article CAS PubMed PubMed Central Google Scholar * Seto E, Yoshida M. Erasers of histone acetylation: the histone
deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a18713. Article Google Scholar * Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3
exhibits decrotonylase activity in vitro. Angew Chem Int Ed. 2012;51:9083–7. Article CAS Google Scholar * Kelly RDW, Chandru A, Watson PJ, Song Y, Blades M, Robertson NS, et al. Histone
deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci Rep. 2018;8:14690–9. Article CAS PubMed PubMed Central Google Scholar * Feldman JL,
Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–6. Article CAS PubMed
PubMed Central Google Scholar * Bao X, Wang Y, Li X, Li X, Liu Z, Yang T, et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach.
Elife. 2014;3:e2999. Article Google Scholar * Jenuwein T. Translating the histone code. Science. 2001;293:1074–80. Article CAS PubMed Google Scholar * Flynn EM, Huang OW, Poy F,
Oppikofer M, Bellon SF, Tang Y, et al. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015;23:1801–14. Article CAS
PubMed Google Scholar * Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell.
2016;62:181–93. Article CAS PubMed PubMed Central Google Scholar * Xiong X, Panchenko T, Yang S, Zhao S, Yan P, Zhang W, et al. Selective recognition of histone crotonylation by double
pHd fingers of MOZ and dpF2. Nat Chem Biol. 2016;12:1111–8. Article CAS PubMed PubMed Central Google Scholar * Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, et al. YEATS2 is a selective
histone crotonylation reader. Cell Res. 2016;26:629–32. Article CAS PubMed PubMed Central Google Scholar * Zhang Q, Zeng L, Zhao C, Ju Y, Konuma T, Zhou M. Structural insights into
histone crotonyl-lysine recognition by the AF9 YEATS domain. Structure. 2016;24:1606–12. Article PubMed PubMed Central CAS Google Scholar * Jiang Y, Chen G, Li X, Liu S, Tian G, Li Y,
et al. Selective targeting of AF9 YEATS domain by cyclopeptide inhibitors with preorganized conformation. J Am Chem Soc. 2020;142:21450–9. Article CAS PubMed Google Scholar * Moustakim
M, Christott T, Monteiro OP, Bennett J, Giroud C, Ward J, et al. Discovery of an MLLT1/3 YEATS domain chemical probe. Angew Chem. 2018;130:16540–5. Article Google Scholar * Heidenreich D,
Moustakim M, Schmidt J, Merk D, Brennan PE, Fedorov O, et al. Structure-based approach toward identification of inhibitory fragments for Eleven-Nineteen-Leukemia Protein (ENL). J Med Chem.
2018;61:10929–34. Article CAS PubMed Google Scholar * Christott T, Bennett J, Coxon C, Monteiro O, Giroud C, Beke V, et al. Discovery of a selective inhibitor for the YEATS domains of
ENL/AF9. SLAS Discov. 2019;24:133–41. CAS PubMed Google Scholar * Asiaban JN, Milosevich N, Chen E, Bishop TR, Wang J, Zhang Y, et al. Cell-based ligand discovery for the ENL YEATS
domain. Acs Chem Biol. 2020;15:895–903. Article CAS PubMed PubMed Central Google Scholar * Li X, Li X, Jiang Y, Liu Z, Cui Y, Fung KY, et al. Structure-guided development of YEATS
domain inhibitors by targeting π–π–π stacking. Nat Chem Biol. 2018;14:1140–9. Article CAS PubMed PubMed Central Google Scholar * Abu-Zhayia ER, Machour FE, Ayoub N. HDAC-dependent
decrease in histone crotonylation during DNA damage. J Mol Cell Biol. 2019;11:804–6. Article PubMed PubMed Central Google Scholar * Liu Y, Li M, Fan M, Song Y, Yu H, Zhi X, et al.
Chromodomain Y-like protein – mediated histone crotonylation regulates stress-induced depressive behaviors. Biol Psychiatry. 2019;8:635–49. Article CAS Google Scholar * Wang Z, Zhao Y, Xu
N, Zhang S, Wang S, Mao Y, et al. NEAT1 regulates neuroglial cell mediating A β clearance via the epigenetic regulation of endocytosis ‑ related genes expression. Cell Mol Life Sci.
2019;15:3005–18. Article CAS Google Scholar * Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res.
2011;21:779–92. Article CAS PubMed PubMed Central Google Scholar * Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, et al. Telomeres acquire embryonic stem cell
characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–54. Article CAS PubMed Google Scholar * Fu H, Tian C, Ye X, Sheng X, Wang H, Liu Y, et al. Dynamics of
telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Rep. 2018;11:70–87. Article CAS Google Scholar * Lv Y, Bu C, Meng J, Ward C, Volpe G, Hu J, et al.
Global profiling of the lysine crotonylome in different pluripotent states. Genom Proteom Bioinform 2021. https://doi.org/10.1016/j.gpb.2021.01.004. * Hakre S, Chavez L, Shirakawa K, Verdin
E. Epigenetic regulation of HIV latency. Curr Opin Hiv Aids. 2011;6:19–24. Article PubMed Google Scholar * Jiang G, Nguyen D, Archin NM, Yukl SA, Méndez-Lagares G, Tang Y, et al. HIV
latency is reversed by ACSS2-driven histone crotonylation. J Clin Investig. 2018;128:1190–8. Article PubMed PubMed Central Google Scholar * Tsilimigras MCB, Fodor A, Jobin C.
Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol. 2017;2:17008–17. Article CAS PubMed PubMed Central Google Scholar * Wan J, Liu H, Ming L. Lysine
crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976–82. Article CAS PubMed Google Scholar * Liu S, Yu H, Liu Y, Liu X, Zhang Y, Bu C, et
al. Chromodomain protein CDYL acts as a Crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol Cell. 2017;67:853–66. Article CAS PubMed Google Scholar * Crespo
M, Damont A, Blanco M, Lastrucci E, Kennani SE, Ialy-Radio C, et al. Multi-omic analysis of gametogenesis reveals a novel signature at the promoters and distal enhancers of active genes.
Nucleic Acids Res. 2020;48:4115–38. Article CAS PubMed PubMed Central Google Scholar * Ruiz-Andres O, Sanchez-Niño MD, Cannata-Ortiz P, Ruiz-Ortega M, Egido J, Ortiz A, et al. Histone
lysine crotonylation during acute kidney injury in mice. Dis Models Mech. 2016;9:633–45. CAS Google Scholar * Tang X, Chen X, Sun X, Xu P, Zhao X, Tong Y, et al. Short-chain enoyl-CoA
hydratase mediates histone crotonylation and contributes to cardiac homeostasis. Circulation. 2021;10:1066–9. Article Google Scholar * Liu K, Yuan C, Li H, Chen K, Lu L, Shen C, et al. A
qualitative proteome-wide lysine crotonylation profiling of papaya (_Carica papaya L._). Sci Rep 2018;8. * Liu S, Xue C, Fang Y, Chen G, Peng X, Zhou Y, et al. Global involvement of lysine
crotonylation in protein modification and transcription regulation in rice. Mol Cell Proteom. 2018;17:1922–36. Article CAS Google Scholar * Xu M, Luo J, Li Y, Shen L, Zhang X, Yu J, et
al. First comprehensive proteomics analysis of lysine crotonylation in leaves of peanut (_Arachis hypogaea L._). Proteomics. 2021;21:2000156. Article CAS Google Scholar * Lu Y, Xu Q, Liu
Y, Yu Y, Cheng Z, Zhao Y, et al. Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol
2018;19:144. * Huang Q, Liao X, Yang X, Luo Y, Lin P, Zeng Q, et al. Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum. Plant
Biotechnol J 2021;19. * Lin P, Bai H, He L, Huang Q, Zeng Q, Pan Y, et al. Proteome-wide and lysine crotonylation profiling reveals the importance of crotonylation in chrysanthemum
(_Dendranthema grandiforum_) under low-temperature. Bmc Genomics 2021;22:51. * Sun J, Qiu C, Qian W, Wang Y, Sun L, Li Y, et al. Ammonium triggered the response mechanism of lysine
crotonylome in tea plants. Bmc Genomics 2019;20. * Gowans GJ, Bridgers JB, Zhang J, Dronamraju R, Burnetti A, King DA, et al. Recognition of histone crotonylation by Taf14 links metabolic
state to gene expression. Mol Cell. 2019;76:909–21. Article CAS PubMed PubMed Central Google Scholar * Peil K, Jürgens H, Luige J, Kristjuhan K, Kristjuhan A. Taf14 is required for the
stabilization of transcription pre-initiation complex in _Saccharomyces cerevisiae_. Epigenetics Chromatin 2020;13. * Sun C, Xu W, Zhao Q, Luo S, Chen X, Li Y, et al. Crotonylation of key
metabolic enzymes regulates carbon catabolite repression in _Streptomyces roseosporus_. Commun Biol 2020;3:192. * Wang Q, Verma J, Vidan N, Wang Y, Tucey TM, Lo TL, et al. The YEATS domain
histone crotonylation readers control virulence-related biology of a major human pathogen. Cell Rep. 2020;31:107528. Article CAS PubMed Google Scholar * Zhou X, Song N, Li D, Li X, Liu
W. Systematic analysis of the lysine crotonylome and multiple posttranslational modification analysis (acetylation, succinylation, and crotonylation) in _Candida albicans_. _Msystems_
2021;6:e01316–20. * Yang Q, Li Y, Apaliya MT, Zheng X, Serwah BNA, Zhang X, et al. The response of _Rhodotorula mucilaginosa_ to patulin based on lysine crotonylation. Front Microbiol
2018;9:2025. * Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating tetrahymena. Proc Natl Acad Sci USA. 1985;23:8048–52.
Article Google Scholar * Xie X, Li X, Qin F, Lin J, Zhang G, Zhao J, et al. Genetically encoded photoaffinity histone marks. J Am Chem Soc. 2017;139:6522–5. Article CAS PubMed Google
Scholar * Bos J, Muir TW. A chemical probe for protein crotonylation. J Am Chem Soc. 2018;140:4757–60. Article CAS PubMed PubMed Central Google Scholar * Xie Y, Yang L, Chen Q, Zhang
J, Feng L, Chen J, et al. Single-step fluorescent probes to detect decrotonylation activity of HDACs through intramolecular reactions. Eur J Med Chem. 2021;212:113120. Article CAS PubMed
Google Scholar * Spinck M, Neumann Staubitz P, Ecke M, Gasper R, Neumann H. Evolved, selective erasers of distinct lysine acylations. Angew Chem Int Ed. 2020;59:11142–9. Article CAS
Google Scholar * Huang G, Zeng W. A discrete hidden Markov model for detecting histone crotonyl lysine sites. MATCH Commun Math Computer Chem. 2015;1:717–30. Google Scholar * Qiu W, Sun B,
Tang H, Huang J, Lin H. Identify and analysis crotonylation sites in histone by using support vector machines. Artif Intell Med. 2017;83:75–81. Article PubMed Google Scholar * Ju Z, He
J. Prediction of lysine crotonylation sites by incorporating the composition of k -spaced amino acid pairs into Chou’s general PseAAC. J Mol Graph Model. 2017;77:200–4. Article CAS PubMed
Google Scholar * Qiu W, Sun B, Xiao X, Xu Z, Jia J, Chou K. iKcr-PseEns: identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics.
2018;110:239–46. Article CAS PubMed Google Scholar * Malebary SJ, Rehman MSU, Khan YD. iCrotoK-PseAAC: identify lysine crotonylation sites by blending position relative statistical
features according to the Chou’s 5-step rule. PloS One. 2019;14:e223993. Article CAS Google Scholar * Liu Y, Yu Z, Chen C, Han Y, Yu B. Prediction of protein crotonylation sites through
LightGBM classifier based on SMOTE and elastic net. Anal Biochem. 2020;609:113903. Article CAS PubMed Google Scholar * Lv H, Dao F, Guan Z, Yang H, Li Y, Lin H. Deep-Kcr: accurate
detection of lysine crotonylation sites using deep learning method. Brief Bioinform. 2020;00:1–10. Google Scholar * Bao C, Song C, Liu Y, Yang Y, Cui Z. Large-scale lysine crotonylation
analysis reveals its potential role in spermiogenesis in the Chinese mitten crab _Eriocheir sinensis_. J Proteom. 2020;226:103891. Article CAS Google Scholar * Zhang N, Yang Z, Liang W,
Liu M. Global proteomic analysis of lysine crotonylation in the plant pathogen _Botrytis cinerea_. Front Microbiol 2020;11:564350. * Kwon OK, Kim SJ, Lee S. First profiling of lysine
crotonylation of myofilament proteins and ribosomal proteins in zebrafish embryos. Sci Rep 2018;8. * Chen W, Tang D, Xu Y, Zou Y, Sui W, Dai Y, et al. Comprehensive analysis of lysine
crotonylation in proteome of maintenance hemodialysis patients. Medicine. 2018;97:e12035. Article CAS PubMed PubMed Central Google Scholar * Lin H, Tang D, Xu Y, Zhang R, Ou M, Zheng F,
et al. Quantitative analysis of protein crotonylation identifies its association with immunoglobulin A nephropathy. Mol Med Rep. 2020;21:1242–50. CAS PubMed PubMed Central Google Scholar
* Liu J, Wu S, Liu S, Sun X, Wang X, Xu P, et al. Global lysine crotonylation profiling of mouse liver. Proteomics. 2020;19–20:e2000049. Article CAS Google Scholar * Li F, Nie L,
Elsheikha HM, Yin F, Zhu X. Lysine crotonylation is widespread on proteins of diverse functions and localizations in Toxoplasma gondii. Parasitol Res 2021;120:1–10. Download references
ACKNOWLEDGEMENTS We thank members of our laboratories for their suggestions and help. This study was supported by the National Key R&D Program of China under grant 2017YFA0506300 (to
K.L.), the National Natural Science Foundation under grant 81902997 (to H.L.) and under grants 31770820 (to K.L.), the Department of Science and Technology of Sichuan Province under grant
2020YJ0047 (to H.L.). AUTHOR INFORMATION Author notes * These authors contributed equally: Gaoyue Jiang, Chunxia Li, Meng Lu. AUTHORS AND AFFILIATIONS * West China Second University
Hospital, State Key Laboratory of Biotherapy, and Key Laboratory of Birth Defects and Related Diseases of Women and Children, Ministry of Education, Sichuan University, 610041, Chengdu,
China Gaoyue Jiang & Huihui Li * Department of Neurosurgery, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University and The Research Units of West China, Chinese
Academy of Medical Sciences, Chengdu, China Chunxia Li, Meng Lu & Kefeng Lu Authors * Gaoyue Jiang View author publications You can also search for this author inPubMed Google Scholar *
Chunxia Li View author publications You can also search for this author inPubMed Google Scholar * Meng Lu View author publications You can also search for this author inPubMed Google Scholar
* Kefeng Lu View author publications You can also search for this author inPubMed Google Scholar * Huihui Li View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS G.J., C.L., and M.L. collected the related references, wrote the manuscript, and constructed the figures; K.L. and H.L. revised the manuscript and supervised this
review. All authors approved the final manuscript and agreed to be responsible for this review. CORRESPONDING AUTHORS Correspondence to Kefeng Lu or Huihui Li. ETHICS DECLARATIONS ETHICS
STATEMENT This review did not require ethical approval. COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. Edited by B. Zhivotovsky SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY
FIGURE 1 SUPPLEMENTARY FIGURE 2 SUPPLEMENTARY FIGURE 3 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which
permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to
the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless
indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or
exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Jiang, G., Li, C., Lu, M. _et al._ Protein lysine crotonylation: past, present, perspective. _Cell Death Dis_ 12, 703 (2021).
https://doi.org/10.1038/s41419-021-03987-z Download citation * Received: 23 February 2021 * Revised: 28 May 2021 * Accepted: 31 May 2021 * Published: 14 July 2021 * DOI:
https://doi.org/10.1038/s41419-021-03987-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative