Lass2 enhances p53 protein stability and nuclear import to suppress liver cancer progression through interaction with mdm2/mdmx
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT LASS2 functions as a tumor suppressor in hepatocellular carcinoma (HCC), the most common type of primary liver cancer, but the underlying mechanism of its action remains largely
unknown. Moreover, details on its role and the downstream mechanisms in Cholangiocarcinoma (CCA) and hepatoblastoma (HB), are rarely reported. Herein, LASS2 overexpression was found to
significantly inhibit proliferation, migration, invasion and induce apoptosis in hepatoma cells with wild-type (HB cell line HepG2) and mutated p53 (HCC cell line HCCLM3 and CCA cell line
HuCCT1). Gene set enrichment analysis determined the enrichment of the differentially expressed genes caused by LASS2 in the p53 signaling pathway. Moreover, the low expression of LASS2 in
HCC and CCA tumor tissues was correlated with the advanced tumor-node-metastasis (TNM) stage, and the protein expression of LASS2 positively correlated with acetylated p53 (Lys373) protein
levels. At least to some extent, LASS2 exerts its tumor-suppressive effects in a p53-dependent manner, in which LASS2 interacts with MDM2/MDMX and causes dual inhibition to disrupt p53
degradation by MDM2/MDMX. In addition, LASS2 induces p53 phosphorylation at ser15 and acetylation at lys373 to promote translocation from cytoplasm to nucleus. These findings provide new
insights into the LASS2-induced tumor suppression mechanism in liver cancer and suggest LASS2 could serve as a potential therapeutic target for liver cancer. SIMILAR CONTENT BEING VIEWED BY
OTHERS ELEVATED EXPRESSION OF WSB2 DEGRADES P53 AND ACTIVATES THE IGFBP3-AKT-MTOR-DEPENDENT PATHWAY TO DRIVE HEPATOCELLULAR CARCINOMA Article Open access 04 January 2024 SALIS
TRANSCRIPTIONALLY REPRESSES IGFBP3/CASPASE-7-MEDIATED APOPTOSIS BY ASSOCIATING WITH STAT5A TO PROMOTE HEPATOCELLULAR CARCINOMA Article Open access 23 July 2022 USP8 POSITIVELY REGULATES
HEPATOCELLULAR CARCINOMA TUMORIGENESIS AND CONFERS FERROPTOSIS RESISTANCE THROUGH Β-CATENIN STABILIZATION Article Open access 13 June 2023 INTRODUCTION Liver cancer, the most common
malignant tumor in the world, ranks sixth in primary cancer incidence and the fourth in cancer-related mortality [1]. In recent decades, the incidence rates of liver cancer have increased in
several countries [2,3,4], with over half of the new cases and deaths occurring in China [5]. Owing to varying risk factors, genetic susceptibility, histopathological typing, and tumor
microenvironment, the heterogeneity of liver cancer considerably limits its early detection [5]. With the rapid progression of the disease, patients are often diagnosed with advanced stages
of liver cancer, thus resulting in an extremely low survival rate. Moreover, owing to the lack of drugs that specifically target the HCC cells, the prognosis is also poor. This highlights
the urgency to develop novel therapeutic interventions. The development and progression of liver cancer are associated with tumor suppressor gene inactivation and oncogene activation. An
in-depth understanding of these complex cellular and molecular networks will provide new perspectives to improve the outcome of patients with liver cancer. LAG1 longevity assurance homolog 2
(LASS2), one of the housekeeping genes of the human genome [6,7,8], is also known as ceramide synthase 2 (CerS2) or tumor metastasis suppressor gene 1 (TMSG1). It is highly expressed in the
liver [8, 9]and kidney [8, 10], particularly in hepatocytes [11]. Depending on the cancer type, the heterogeneous expressions of LASS2 are associated with either cancer progression or
suppression. Most studies have indicated an interrelationship between low LASS2 mRNA or protein expression and the degree of poor prognosis and invasion in diseases including in liver cancer
[12, 13], prostate cancer [14, 15], bladder cancer [16], breast cancer [17, 18]. Thus, LASS2 has been recognized as a tumor suppressor gene. However, a few studies have found that the
increased mRNA or protein levels of LASS2 were observed in metastatic cell lines or tumor specimens as compared to non-metastatic cell lines or paracancerous tissue [19, 20]. These increased
mRNA or protein levels of LASS2 are correlated with FIGO staging in ovarian cancer patients [20], implicating that LASS2 may act as an oncogene. In a previous study in HepG2 hepatoblastoma
cells (HB) [21], the overexpression of LASS2 inhibited proliferation and induce apoptosis. Studies have reported that low LASS2 expression is associated with a poor prognosis for patients
with hepatocellular carcinoma (HCC) [22] and suggested that the combination of LASS2 and TGF-β1 [22] or ASGR1 [23] may assist in predicting the prognosis. All these findings indicate that
LASS2 might be independently used as a prognostic marker for HCC patients [23]. To date, the underlying molecular mechanisms of LASS2 reported for HCC regulation remain poorly understood,
and its role and mechanisms in other subtypes of liver cancer (such as CCA and HB) are also poorly reported. Therefore, substantial research is necessary before the exact molecular
mechanisms of LASS2 tumor-suppressive action is elucidated, and the research in this field needs to be expanded. p53, a well-known tumor suppressor gene involved in cell cycle control,
apoptosis, cell differentiation [24,25,26], and epithelial-mesenchymal transition (EMT) [27], is one of the most mutated genes in HCC and a potent regulator of metastasis [27]. It is
involved in regulating the mitochondrial apoptosis pathway [28], The results of our previous study showed that the overexpression of LASS2 over-elicited mitochondrial apoptosis in HepG2
cells [21], but the relation between LASS2 and p53 remains unclear. Herein, the relationship between LASS2 and p53 is investigated, its prognostic value in liver cancer is assessed, and the
underlying molecular mechanisms are investigated. RESULTS OVEREXPRESSION OF LASS2 SUPPRESSES PROLIFERATION, INVASION, AND MIGRATION, AND PROMOTES APOPTOSIS IN LIVER CANCER CELL LINES
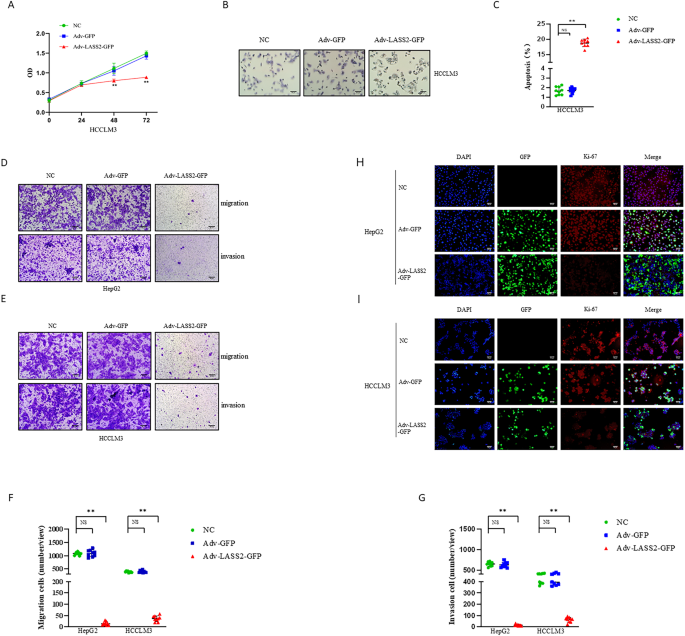
According to a previous report, the overexpression of LASS2 inhibits proliferation and induces apoptosis in HepG2 HB cells [21]. To investigate whether LASS2 also alters these biological
characteristics of HCCLM3 HCC cells and HuCCT1 CCA cells, the CCK-8 and dUTP TUNEL assay were performed. The results consistently suggested that HCCLM3 and HuCCT1 cells with LASS2
overexpression displayed lower proliferation rates and could induce higher apoptosis cells compared with that in the negative or vector-control cells (all _P_ < 0.05, Fig. 1A–C and Fig.
S1A–C). The results from the Ki-67 immunofluorescence staining assay further indicated that LASS2 overexpression decreased the proliferation ability of HepG2, HCCLM3, and HuCCT1 cells (Fig.
1H, I and Fig. S1G). The findings of the transwell migration and matrigel invasion assays revealed that LASS2 overexpression resulted in a lower migration and invasion capability compared
with the negative control or vector-control cells in the liver cancer cell lines (all _P_ < 0.01, Fig. 1D–G and Fig. S1D–F). DOWN-REGULATED EXPRESSION OF LASS2 PREDICTED A POOR PROGNOSIS
OF LIVER CANCER IN THE TEST COHORT To investigate the role of LASS2 in liver cancer progression, 90 liver cancer specimens (the test cohort consists of 60 HCC patients and 30 human CCA
patients) were collected and stained using immunohistochemistry (IHC) to assess LASS2 protein expression. The results of IHC staining analysis revealed that LASS2 protein is relatively
abundant in the early-stage disease (TNM stages I or II) and scarce in later-stage liver cancer (TNM stages III and IV) (Fig. 2A). Statistical analyses, based on the semi-quantification of
the IHC staining (ID score), indicated that LASS2 expression level was negatively correlated to the TNM stage (_P_ = 0.013, Table 1) (TNM stages II vs. I: _P_ = 0.011; TNM stages III–IV vs.
I: _P_ < 0.001, Fig. 2B) and tumor size (_P_ = 0.029, Table 1), and lower LASS2 expression in CCA tumor samples compared to HCC tissue by violin plot analysis (_P_ = 0.0052, Fig. 2C). As
shown in Table 1, sex, age and the concentration of AFP in the patient serum were not significantly correlated to LASS2 expression levels. Using the KM plotter database including 118 tumor
samples in HCC patients (_P_ = 0.018, Fig. 2D), Kaplan–Meier survival curves revealed that liver cancer patients with lower LASS2 expression had significantly reduced overall survival. LASS2
INDUCES THE PHOSPHORYLATION AND ACETYLATION OF P53 AND ACTIVATES THE P53 SIGNALING PATHWAY As shown by the GeneSet enrichment analysis (GSEA), the LASS2-induced differentially expressed
genes were enriched in the p53 signaling pathway (Gene Ontology Biological Process, GOBP analysis) (_P_ = 0.006, FDR _q_ = 0.591, Fig. 2E). An analysis of TCGA-LIHC (Liver hepatocellular
carcinoma) dataset from GTBAdb online tools indicated the significant and positive correlation of LASS2 (CERS2) expression levels with TP53 mRNA expression (_r_ = 0.138, _P_ = 0.008, Fig.
2F). Therefore, it was further investigated whether LASS2 affects the signaling pathway of p53. The western blotting results (Fig. 2G) revealed that LASS2 overexpression did not alter total
p53 levels. Interestingly, the enhanced phosphorylation of p53 (at ser15) and acetylation of lys373 (acetyl lys373), and increased p53 directly targeting genes such as p21 were observed in
LASS2-overexpressing TP53 wild-type cell line HepG2 cells (Fig. 2G). Similarly, in LASS2-overexpressing TP53 mutant cell lines (HCCLM3 and HuCCT1), p53 phosphorylation and acetylation still
occurred, and the p21 protein level was also upregulated (Fig. 2H and Fig. S2A). The correlation between LASS2 and acetylation of p53 at lys373 (acetyl lys373) was confirmed using 60 HCC and
30 CCA. Similar results were observed in clinical liver cancer sections by IHC, and the LASS2 protein levels were also found to be positively correlated with acetylation of p53 (lys373)
expression (_r_ = 0.3151, _P_ = 0.003, Fig. 2I, J). Collectively, these results indicate that the overexpression of LASS2 activates the p53 signaling pathway. LASS2 PROMOTES P53-MEDIATED
MITOCHONDRIAL APOPTOSIS IN LIVER CANCER CELLS To determine the role of p53 in mitochondrial apoptosis, the proteins involved in p53-mediated apoptosis were further assessed by western blot.
The western blotting results also confirmed the close association of LASS2 with mitochondrial apoptosis in HepG2, HCCLM3, and HuCCT1 cell lines. The overexpression of LASS2 reduced Bcl-2
expression while upregulating the protein levels of Bax, Cyto-c, Cleaved-caspase-3, and Cleaved-caspase-9 (all _P_ < 0.05, Fig. 3A, B and Fig. S2B). As PUMA is a downstream target gene of
p53 and promotes apoptosis through p53-dependent and p53-independent pathways, and given the crucial role for PUMA and Cyto-c, Bcl-2 mediated mitochondrial apoptotic pathway, we examined
whether these pathways may also promote apoptosis in response to p53-inhibition by pifithrin-α (PFT-α). As shown in Fig. 3C, D and Fig. S2C, after PFT-α treatment, overexpression of LASS2
attenuated PUMA and Cyto-c, Bcl-2 mediated mitochondrial apoptosis, downstream target of p53. LASS2 IMPEDES EMT AND ECM IN LIVER CANCER CELL LINES THROUGH THE P53 SIGNALING PATHWAY To
further study the mechanism by which LASS2 regulates cell migration and invasion, the protein levels of EMT markers in LASS2 overexpressed cell lines and control cell lines were determined.
The results showed that LASS2 overexpression increased the protein expression of epithelial cell markers (E-cadherin), while decreasing the expression of mesenchymal cell markers
(N-cadherin, vimentin) and EMT transcriptional drivers (Snail, Slug) in HepG2, HCCLM3, and HuCCT1 cells (all _P_ < 0.05, Fig. 4A, B and Fig. S2D). Two matrix metalloproteinases (MMPs)
MMP2 and MMP9 are the endopeptidases involved in ECM remodeling, EMT, and tumor cell invasion [29,30,31]. Consistent with an EMT phenotype, HepG2, HCCLM3, and HuCCT1 cells with LASS2
overexpression showed the diminished degradative capabilities of ECM, as indicated by their reduced MMP2 and MMP9 expressions (all _P_ < 0.05, Fig. 4C, D and Fig. S2E). Importantly, the
effect of LASS2 overexpression on the expression of p53 downstream EMT targets (Slug, N-cadherin, and vimentin) in _WT_ p53 and mutant p53 hepatoma cell lines (_P_ < 0.05, Fig. 4E, F and
Fig. S2F) was also found to be attenuated by the p53 inhibitor PFT-α. LASS2 DIRECTLY INTERACTS WITH MDMX/MDM2 The capability of LASS2 to significantly influence liver cancer cell line
phenotype has been demonstrated in the current study. However, the underlying molecular mechanism remains unknown to date. Further research is needed to clarify the regulatory mechanisms of
LASS2 in the inhibition of liver cancer progression, the possible LASS2 interaction proteins were identified by co-immunoprecipitation (co-IP)-coupled Liquid Chromatography-Mass Spectrometry
(LC/MS) in Hepa1-6 hepatoma cells. The higher number of differential bands specific to LASS2-GFP in the coomassie blue stained SDS-PAGE gel was observed as compared to the GFP control, as
shown in Fig. 5A and Fig. S3A. Protein interaction analysis (PIP) demonstrated that the p53 signaling pathway-related proteins might interact with LASS2, including APAF1, MDM2, MDMX, CDK1,
PCNA, EPHA2, TJP1, HNRNPK, and AIFM1 (Fig. 5B). Notably, both MDM2 and MDMX are the critical negative regulators of p53 function [32, 33]. To determine the direct interaction between LASS2
and MDM2 or MDMX, their interactions were studied by co-IP using GFP Nanoselector beads for immunoprecipitation in TP53 wild-type HepG2 and TP53 mutant HCCLM3 and HuCCT1 cell lines, followed
by western blot with anti-GFP, anti-LASS2, anti-MDM2, and anti-MDMX. Co-IP western blot results confirmed the presence of interactions between LASS2 and MDM2 or MDMX (Fig. 5C and Fig. S3B).
Proximity Ligation Assays (PLAs) further confirmed these findings (Fig. 5D–G). Significant PLA signals were detected in HepG2 (WT p53) and HCCLM3 (mutant p53) cells, and the PLA interaction
signals of LASS2 with MDM2 or MDMX were primarily localized in the nucleus of HepG2 (WT p53) but in the cytoplasm of HCCLM3 (mutant p53) cells. Therefore, protein–protein docking studies
were performed to gain insights into the nature of molecular interactions between LASS2 with MDM2 or MDMX. The docking results were obtained by selecting the lowest energy model from the ten
models with low binding energies according to their Weighted Scores. The contact lists between LASS2 with MDM2 or MDMX are shown in Table S1–3. Next, the protein interaction binding modes
and sites between these proteins were predicted using PyMol software. The results of docking studies in the LASS2-MDM2 or MDMX binding models are shown in Fig. 5H, I and Tables S2, 3.
Domains are considered to be the structural and functional units of a protein, LASS2 protein has distinct Homeobox-like domains (amino acids 71–128), Tram-Lag-CLN8 (TLC) domains (amino acids
131–332), and Lag1p motifs. By using ZDOCK 3.0.2 to predict functional protein domains, LASS2 was found to probably interact with MDM2 or MDMX through its Homeobox-like domains and TLC
domain (Fig. 5J). Interestingly, the binding sites of LASS2 and MDM2 or MDMX overlap with the previously reported [34] binding sites of p53 and MDM2 or MDMX (Fig. 5J). LASS2 INHIBITS
MDM2/MDMX EXPRESSION AND ENHANCES P53 STABILITY IN LIVER CANCER CELLS As shown in Fig. 6A, B, using the Kaplan–Meier Plotter database (MDM2 DFS: Hazard ratio [HR] = 2.1, _P_ = 0.0018; MDMX
DFS: HR = 1.8, _P_ = 0.012), multivariable analysis for Disease Free Survival (DFS) determined that high MDM2 or MDMX expression is associated with shorter DFS in liver cancer patients. To
further elucidate the regulatory relationship between LASS2 and MDM2 or MDMX in the process of liver cancer propagation, an online TNMplot database was used to conduct a computational
analysis of these relationships in LIHC. The results demonstrated that LASS2/CERS2 transcript levels were negatively correlated with MDM2 or MDMX expression (MDM2: _r_ = −0.20, _P_ <
0.01; MDMX: _r_ = −0.13, _P_ < 0.01, Fig. 6C, D). In addition, LASS2 overexpression was found to inhibit the mRNA and protein expression levels of MDM2 or MDMX in HepG2, HCCLM3, and
HuCCT1 cells (all _P_ < 0.01, Fig. 6E–H and Fig. S3C–E), respectively. As the dual inhibition of MDMX and MDM2 is required for the full release of dormant p53 [33], dual inhibition is a
potential pathway for p53 activation [35]. Dual luciferase reporter assays were performed to investigate whether LASS2 regulates the p53 signaling pathway. A remarkable increase in the
transcriptional activity of p53 was observed by LASS2 overexpression (Fig. 6I). Furthermore, the p53 protein levels in the nucleus and cytoplasm were studied. As shown in Fig. 6J, K and Fig.
S3F, the overexpression of LASS2 stimulated the nuclear translocation of p53 in TP53 wild-type cell lines and TP53 mutant cell lines. Collectively, these findings further indicated that
LASS2 activates the p53 signaling pathways and enhances p53 stability in HepG2, HCCLM3, and HuCCT1 cells. DISCUSSION HCC, CCA, and HB collectively represent the major forms of liver
malignancies with high recurrence and metastasis rates and are malignancies with poor prognosis [36,37,38,39]. Abnormal gene expression is significantly associated with the occurrence and
inadequate prognosis of these three liver cancer subtypes [40]. Although low LASS2 expression in tumor tissues of HCC patients is associated with prognosis [22, 23], a limited understanding
of its expression in other liver cancer subtypes and the underlying molecular mechanisms is available. In the current work, functional studies revealed that LASS2 inhibited the
proliferation, apoptosis, invasion, and migration of different subtypes of liver cancer cell lines. Next, LASS2 expression was found to be down-regulated in patients with HCC and CCA, which
correlated with their poor prognosis. Bioinformatics predictions indicated that high LASS2 expression was associated with the p53 signaling pathway, which was experimentally verified. The
p53 protein is a sequence-specific DNA-binding transcription factor encoded by the TP53 gene and is known as the “guardian of the genome” [41]. p53 activity is usually regulated at the
protein level by post-translational modifications (PTM) [42]. Under stress-free conditions, p53 protein levels are usually maintained low by continuous degradation. In stressed cells, the
p53 protein is subject to various post-translational modifications that affect the expression of p53 target genes, including phosphorylation and acetylation [42]. The data from the current
study clearly show that LASS2 overexpression facilitates the phosphorylation of p53 at ser15 and acetylation of p53 at lys373 and increases the protein expression of the key downstream
target gene p21 in TP53 wild-type HepG2 and mutant HCCLM3 and HuCCT1 cell lines. Additionally, LASS2 overexpression promoted the transcriptional activity of p53, as revealed by the dual
luciferase reporter system. Therefore, LASS2 is suggested to regulate p53 activation through a two-pronged approach. Subsequent Cox proportional risk regression model analysis obtained from
TCGA-LIHC further demonstrated the positive correlation of LASS2 expression with TP53 mRNA expression levels. Owing to the unstable expression and rapid degradation of wild-type p53, it
cannot be detected by the IHC method [43]. In addition, acetylation is critical for p53 because it enhances p53 protein stability [44], binds to other proteins, and is required for its
checkpoint response to stress [45]. Therefore, the correlation between LASS2 and acetylated p53 was analyzed in clinical samples and the expression of LASS2 was positively associated with
the level of acetylated p53 (lys373) protein expression. Collectively, the results of the present study support the association between LASS2 and activation of the p53 signaling pathway
(Fig. 7). Further, LASS2 is found to exhibit a direct interaction with MDM2 and MDMX, and LASS2 overexpression is found to suppress the expressions of MDM2 and MDMX. MDM2 is a ubiquitin
ligase that targets and degrades p53 through the ubiquitin-proteasome system [33]. MDMX, also known as MDM4, is a structural homolog of MDM2, but it lacks intrinsic E3 ubiquitin ligase
binding activity. Moreover, it primarily inhibits p53 transcriptional activity but does not affect its protein stability [46, 47]. Several groups have reported that MDM2 and MDMX play
indispensable and non-overlapping roles in inhibiting the normal function of p53, and dual inhibition by MDM2 and MDMX is essential for the full release of dormant p53 and is a potential
pathway for p53 activation [33, 48, 49]. Given that the possible binding domains of LASS2 and MDM2 or MDMX (protein–protein docking in this study) share the same region as the binding
domains of p53 and MDM2 or MDMX, it can be speculated that LASS2 may inhibit p53 degradation by competitively inhibiting the binding of MDM2 or MDMX to p53. The activated tumor suppressor
p53 can regulate the expression of dozens of target genes and different physiological processes [50]. The phosphorylation of p53 is a key modification that directs its regulation of
apoptotic cell death, where phosphorylation at the ser15 site stabilizes p53 by protecting it from MDM2 [42]. The acetylation of the C-terminus of p53 (including lys53, lys370, lys372,
lys373, lys381, and lys382) facilitates its binding to target gene sites to activate downstream pathways, and acetylation of these sites prevents the degradation of p53 and induces its
accumulation [42, 45, 51, 52]. This study demonstrates that LASS2 overexpression contributes to the stabilization and accumulation of p53 protein in the nucleus of wild-type and mutant
hepatoma cell lines, enhances the role of p53 in the regulation of downstream target gene activation or inhibition, and is involved in causing apoptosis (Bcl-2, Bax, Cyto-c, Cleaved
caspase-3 and 9), EMT (Snail, Slug, N-cadherin, E-cadherin and vimentin), and ECM (MMP2 and MMP9) genes. Notably, HCCLM3 and HuCCT1 have mutant p53 status, but both express p53 (compared
with HepG2, primarily expressed in the cytoplasm), and it was speculated whether LASS2 depends on p53 for its oncogenic effects when LASS2 overexpression exhibits the same biological role in
these three cell lines with different p53 status. Unexpectedly, the p53 inhibitor pifithrin-α attenuated the effect of LASS2 overexpression on p53 downstream mitochondrial apoptosis target
gene (PUMA), Bcl-2, Cyto-c, and also on EMT target genes (Slug), N-cadherin, and vimentin. The finding of this study suggests that LASS2 exerts a tumor suppressive function, at least in
part, through the p53 signaling pathway in both wild-type and these two p53 mutant types of cell lines. In summary, the results of the current study demonstrate for the first time that LASS2
exerts a tumor-suppressive effects by interacting with MDM2 and MDMX and promoting p53 ser15 phosphorylation and lys373 acetylation in the different subtypes of hepatoma cells that harbor
wild-type and mutant p53, thereby stabilizing p53. As we all know, p53 activation is an attractive strategy for anticancer therapy. These findings highlight the importance of the
LASS2-MDM2/MDMX-p53 signaling axis in the regulation of liver cancer and may elucidate the mechanisms of mutant p53 reactivation, providing possible new targets for clinical treatment.
MATERIALS AND METHODS CELL CULTURE AND TRANSFECTIONS Human HCC cell line HCCLM3 (mutant p53) and human Cholangiocarcinoma (CCA) cell line HuCCT1 (mutant p53, R175H) were obtained from Feng
Hui Biotechnology company (Changsha, Hunan, China). Human HB cell line HepG2 (wild-type p53) and mouse hepatoma cell line Hepa1-6 were kindly provided by Stem Cell Bank, Chinese Academy of
Sciences (Shanghai, China). All cell lines were routinely cultured in DMEM medium (Gibco) or RPMI-1640 (HuCCT1) (Gibco) containing 10% (v/v) fetal bovine serum (FBS) and 1%
penicillin/streptomycin at 37 °C with 5% CO2. The recombinant human adenovirus vector overexpressing LASS2 (Adv-_h_LASS2-GFP), the recombinant mouse adenovirus-expressing LASS2 vector
(Adv-_m_LASS2-GFP), and the control adenovirus vector (Adv-GFP) were constructed as described in a previously reported study [21]. HCCLM3, HuCCT1, and HepG2 cells were transfected with
Adv-GFP or Adv-_h_LASS2-GFP for 48 h, or the cells were treated with PFT-α (20 μM, MCE) for 24 h and then transfected for 48 h, respectively. Next, the cells were separately harvested
according to the requirement of each experiment, and all assays were independently performed in triplicates. CELL VIABILITY ASSAY A CCK-8 assay (Solarbio, Beijing, China) was performed to
determine cellular proliferation. A total of 6000 cells were seeded into 96-well plates (Corning). The plate was incubated overnight with 5% CO2 at 37 °C. At 24, 48, and 72 h
post-transfections with Adv-GFP or Adv-_h_LASS2-GFP, CCK-8 reagent was added into each well. Using a microplate reader (Thermo Multiskan, USA), the optical density of the solution
measurement at 450 nm was performed after incubation for 2 h. CELL MIGRATION AND INVASION ASSAY Transwell chambers (8 μm pore size, 6.5 mm diameter, Corning Costar) were used to study cell
migration and invasion abilities. A total of 5 × 104 cells/mL were added to the upper chamber with serum-free medium. Then, a DMEM medium containing 20% FBS was placed in every well of the
lower chamber. After incubating for 48 h, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet ammonium oxalate solution (Solarbio, China), and the remaining
cells on the upper surface were wiped with cotton balls. Finally, under a microscope (Nikon, Japan), the stained cells that had migrated through the transwell polycarbonate membrane were
observed. Diluted Matrigel (at a ratio: of 1:8) was used in the invasion assay. TERMINAL DEOXYNUCLEOTIDYL TRANSFERASE DUTP NICK END LABELING (TUNEL) ASSAY TUNEL, a terminal deoxynucleotidyl
transferase-dUTP nick end labeling assay (KeyGen Biotech, China), was used to detect cell apoptosis. Briefly, cells are fixed, permeabilized, labeled, ligated, and stained, and then the
number of brown-stained cell nuclei were counted under a microscope (Olympus, Japan). IMMUNOFLUORESCENCE (IF) STAINING After the cells were fixed and permeabilized, the cells were blocked
with 10% goat serum and incubated overnight at 4 °C with a primary antibody against Ki-67 (Proteintech, 273099-1-AP, 1:1000). The cells were incubated with CoraLite594-conjugated goat
anti-rabbit IgG (Proteintech, SA0013-4, 1:100). For the final step, the cells were incubated with Hoechst 33342 (CST, 4082S, 1:1000) for 15 min. Dark conditions were maintained and the cells
were observed under a fluorescent microscope (Olympus, Japan). HUMAN CLINICAL SAMPLES Ninety formalin-fixed, paraffin-embedded tissues were obtained from primary liver cancer patients who
underwent surgery and were untreated with radiotherapy/chemotherapy in the Affiliated Hospital of ZunYi Medical University (Guizhou, China). All samples were acquired after obtaining patient
consent and approval from the Ethics Committee of the Affiliated Hospital of ZunYi Medical University (Ethics approval number: KLL-2019-020). As shown in Table 1, the detailed clinical data
of patients are provided. IMMUNOHISTOCHEMISTRY (IHC) Sixty human HCC and 30 human CCA tissue paraffin sections (4-µm thick) were deparaffinized and IHC was carried out as described
previously [53]. The complete list of primary and secondary antibodies and related information is provided in Table S1. The score for staining was independently assessed by two experienced
and blinded observers based on the staining intensity and percentage of positive cells. The staining intensity was scored as follows: cells with no positive staining were defined as 0
points; 1 point for light yellow; 2 points for yellow or light brown; cells with dark brown staining were defined as 3 points. The percentage of positive cells was scored as follows: cells
with less than 5% staining were scored as 0 points; cells with 5–25% staining were scored as 1 point; the score of 26–50% stained cells was 2 points; 51–75% positively staining cells were
scored as 3 points; and that with 76–100% were scored as 4 points. These two types of points were multiplied to generate overall scores (ID scores). The sum of the average integrated optical
density (IOD) of each sample was calculated using Image J software. REAL-TIME PCR AND WESTERN BLOTTING Following the total RNA extraction with RNAiso Plus reagent (Takara, Japan), and the
total protein fractions of HCCLM3, HuCCT1, and HepG2 cells were extracted with a protein extraction kit (KeyGen Biotech, China). A nuclear and cytoplasmic protein extraction kit (APPLYGEN,
China) was used to extract nuclear and cytoplasmic proteins of the liver cancer cell lines for detecting p53 protein. Protein concentration was measured using the BCA reagent kit (Epizyme
Biotech) according to manufacturer’s instructions. All these procedures were performed as previously described [54]. The primers and antibodies are listed in Tables S4 and S5.
CO-IMMUNOPRECIPITATION (CO-IP) LC-MS/IMMUNOBLOTTING (IB) Hepa1-6 hepatoma cells were transfected with Adv-GFP or Adv-_m_LASS2-GFP for 48 h, and then the cells were lysed with ice-cold IP
lysis/wash buffer (Thermo Scientific Pierce, USA) after washing twice with PBS. As instructed by the manufacturer (GFP Nanoselector Agarose one-step immunoprecipitations kit, NBbiolab,
China), co-IP experiments were performed as described previously [55]. Briefly, to capture GFP-fusion proteins, lysates were incubated for 1 h at 4 °C with 25 μL equilibrated GFP
Nanoselector beads. Thereafter, the beads were washed with wash buffer, resuspended in 2 × SDS-sample buffer, and boiled for 10 min at 95 °C to dissociate the immuno-complexes from the
beads. Using SDS-PAGE, proteins were separated, and stained with Coomassie Blue (Epizyme, China), and then the stained SDS gel portions were excised, destained, dehydrated, dried, and
digested with trypsin. Subsequently, freeze-dried tryptic peptides were subjected to LC-MS analysis. An online tool called STRING (_ver_. 11.5) (https://cn.string-db.org/) was used to
identify the potential interaction networks and functional enrichments of these interactions of proteins (gene ontology (GO) annotation, KEGG pathway). The Co-IP samples of HCCLM3, HuCCT1,
and HepG2 cells transiently transfected with Adv-GFP and Adv-_h_LASS2-GFP were analyzed by western blotting to determine the interaction between LASS2 and MDMX or MDM2. PROXIMITY LIGATION
ASSAY A PLA kit (Duolink, Sigma Aldrich) was used to detect LASS2 and MDM2 or MDMX interactions. In brief, after the cells were fixed and permeabilized, they were permeabilized with 0.1%
TRITON X-100 and incubated with Duolink® Closure Solution. Anti-LASS2 (1:500, sc-390745, Santa cruz, American) and anti-MDM2 (1:200, ER1902-14, HuaBio, China) or MDMX (1:500, 17914-1-AP,
Proteintech, China) were mixed and diluted in Duolink® antibody diluent and incubated overnight at 4 °C. The PLUS and MINUS PLA probes were mixed and diluted in Duolink® antibody diluent
(dilution ratio 1:5) and incubated for 1 h at 37 °C in a preheated humidity chamber. After ligation, amplification, and washing, the slices were sealed with Duolink® in-situ sealer
containing DAPI, waited for 15 min to acquire images under a confocal microscope (Leica SP8, Germany), and analyzed with the confocal software. PROTEIN–PROTEIN DOCKING STUDIES The amino acid
sequences of LASS2 and interacting proteins MDMX or MDM2 were downloaded from the RCSB PDB database (https://www.rcsb.org/). To create the 3D models of these proteins, homology modeling was
performed using SWISS-MODEL (https://swissmodel.expasy.org/). Subsequently, a set of the highest predictive-performing models and their modeling conditions were selected. Protein–protein
docking was performed using the web server ClusPro 2.0 (https://cluspro.org) for these two protein structures to predict their binding interaction. Based on the lowest Weighted Score, the
docking structure was visualized with the PyMol software (version 2.5, available at http://www.pymol.org). BIOINFORMATICS The transcriptome data and corresponding clinical information of HCC
patients were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/), including 374 tumor and 50 normal liver samples. The “limma” R package was used to analyze the
differential expression of specific genes between cancer and normal liver tissues, and the relationship between clinical features and gene expression was also evaluated by R software. GSEA
was performed to investigate the functions of LASS2 based on the TCGA database with the “Clusterprofile” package. _P_ < 0.05 was considered to be statistically significant enriched
function annotations in Gene Ontology Biological Process (GOBP) pathways. Gene correlation analysis was performed through the online tools GTBAdb analysis (https://www.gtbadb.com/) or TNM
plot analysis (https://www.tnmplot.com/). According to the Kaplan–Meier Plotter database (http://kmplot.com/analysis/), survival curves were plotted and analyzed using log-rank tests. A
published study consisting of RNA-seq data and clinical information of 118 HCC patients available at the KM plotter was used to assess overall survival against the LASS2 mRNA data.
DUAL-LUCIFERASE REPORTER ASSAY The HepG2 (wt p53) and HuCCT1(mutant p53) cells stably expressing doxycycline-inducible constructs of LASS2 were pretreated with doxycycline for 48 h before
seeding, and then were plated in 24-well plates at 1.5 × 105 cells per well before the transfection of luciferase reporter plasmids. PG13-luc luciferase reporter and pRL-CMV plasmid (Renilla
luciferase, Promega) were cotransfected into hepatoma cells as mentioned above. As directed by the manufacturer, the activity of luciferase was determined with a dual luciferase reporter
assay system (Promega). STATISTICAL ANALYSIS All data analyses were performed using GraphPad Prism 8.0. Differences in categorical variables were assessed by χ2 tests, and differences in
continuous variables were measured by applying one-way ANOVA. Based on Pearson’s correlation analysis, the relationship between the variables was determined. _P_-values < 0.05 were
considered to be statistically significant (*_P_ < 0.05; **_P_ < 0.01; ***_P_ < 0.001); _P_ > 0.05 was considered non-significant (NS). All bioinformatics analyses of data
downloaded from the TCGA database were performed using statistical software R (version 4.0.3). DATA AVAILABILITY All data used or analyzed during this study are included in this article and
available from the corresponding author upon reasonable request. REFERENCES * Li X, Ramadori P, Pfister D, Seehawer M, Zender L, Heikenwalder M. The immunological and metabolic landscape in
primary and metastatic liver cancer. Nat Rev Cancer. 2021;21:541–57. Article CAS PubMed Google Scholar * McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma.
Hepatology. 2021;73:4–13. Article CAS PubMed Google Scholar * Liu Z, Jiang Y, Yuan H, Fang Q, Cai N, Suo C, et al. The trends in incidence of primary liver cancer caused by specific
etiologies: Results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J Hepatol. 2019;70:674–83. Article PubMed Google Scholar * Cao M, Li H, Sun
D, Chen W. Cancer burden of major cancers in China: a need for sustainable actions. Cancer Commun 2020;40:205–10. Article Google Scholar * Fu J, Wang H. Precision diagnosis and treatment
of liver cancer in China. Cancer Lett. 2018;412:283–8. Article CAS PubMed Google Scholar * Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, Grösch S. The enigma of ceramide synthase
regulation in mammalian cells. Prog Lipid Res. 2016;63:93–119. Article CAS PubMed Google Scholar * Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH Jr, et al.
Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem. 2008;283:5677–84. Article CAS PubMed Google
Scholar * Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–71. Article CAS PubMed Central
PubMed Google Scholar * Lyn-Cook LE Jr, Lawton M, Tong M, Silbermann E, Longato L, Jiao P, et al. Hepatic ceramide may mediate brain insulin resistance and neurodegeneration in type 2
diabetes and non-alcoholic steatohepatitis. J Alzheimers Dis. 2009;16:715–29. Article PubMed Central PubMed Google Scholar * Marsching C, Rabionet M, Mathow D, Jennemann R, Kremser C,
Porubsky S, et al. Renal sulfatides: sphingoid base-dependent localization and region-specific compensation of CerS2-dysfunction. J Lipid Res. 2014;55:2354–69. Article CAS PubMed Central
PubMed Google Scholar * Kremser C, Klemm AL, van Uelft M, Imgrund S, Ginkel C, Hartmann D, et al. Cell-type-specific expression pattern of ceramide synthase 2 protein in mouse tissues.
Histochem Cell Biol. 2013;140:533–47. Article CAS PubMed Google Scholar * Tang N, Jin J, Deng Y, Ke RH, Shen QJ, Fan SH, et al. LASS2 interacts with V-ATPase and inhibits cell growth of
hepatocellular carcinoma. Sheng Li Xue Bao. 2010;62:196–202. CAS PubMed Google Scholar * Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, et al. Adult ceramide synthase
2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J Biol Chem. 2009;284:33549–60. Article CAS PubMed Central PubMed Google Scholar
* Xu X, You J, Pei F. Silencing of a novel tumor metastasis suppressor gene LASS2/TMSG1 promotes invasion of prostate cancer cell in vitro through increase of vacuolar ATPase activity. J
Cell Biochem. 2012;113:2356–63. Article CAS PubMed Google Scholar * Huang L, Luan T, Chen Y, Bao X, Huang Y, Fu S, et al. LASS2 regulates invasion and chemoresistance via ERK/Drp1
modulated mitochondrial dynamics in bladder cancer cells. J Cancer. 2018;9:1017–24. Article PubMed Central PubMed Google Scholar * Aldoghachi AF, Baharudin A, Ahmad U, Chan SC, Ong TA,
Yunus R, et al. Evaluation of CERS2 gene as a potential biomarker for bladder cancer. Dis Markers. 2019;2019:3875147. Article PubMed Central PubMed Google Scholar * Mei F, You J, Liu B,
Zhang M, Liu J, Zhang B, et al. LASS2/TMSG1 inhibits growth and invasion of breast cancer cell in vitro through regulation of vacuolar ATPase activity. Tumour Biol. 2015;36:2831–44. Article
CAS PubMed Google Scholar * Fan S, Niu Y, Tan N, Wu Z, Wang Y, You H, et al. LASS2 enhances chemosensitivity of breast cancer by counteracting acidic tumor microenvironment through
inhibiting activity of V-ATPase proton pump. Oncogene. 2013;32:1682–90. Article CAS PubMed Google Scholar * Ma C, Liu Y, Zheng J, Fang W, You J, Wang J, et al. Identification of tumor
metastasis related gene TMSG-1 by mRNA differential display. Sci China C Life Sci. 2002;45:553–60. Article CAS PubMed Google Scholar * Sheng N, Wang Y, Xie Y, Chen S, Lu J, Zhang Z, et
al. High expression of LASS2 is associated with unfavorable prognosis in patients with ovarian cancer. J Cell Physiol. 2019;234:13001–13. Article CAS PubMed Google Scholar * Yang Y, Yang
X, Li L, Yang G, Ouyang X, Xiang J, et al. LASS2 inhibits proliferation and induces apoptosis in HepG2 cells by affecting mitochondrial dynamics, the cell cycle and the nuclear factor‑κB
pathways. Oncol Rep. 2019;41:3005–14. CAS PubMed Google Scholar * Ruan H, Wang T, Yang C, Jin G, Gu D, Deng X, et al. Co-expression of LASS2 and TGF-β1 predicts poor prognosis in
hepatocellular carcinoma. Sci Rep. 2016;6:32421. Article CAS PubMed Central PubMed Google Scholar * Gu D, Jin H, Jin G, Wang C, Wang N, Hu F, et al. The asialoglycoprotein receptor
suppresses the metastasis of hepatocellular carcinoma via LASS2-mediated inhibition of V-ATPase activity. Cancer Lett. 2016;379:107–16. Article CAS PubMed Google Scholar * Zeng K, Chen
X, Hu X, Liu X, Xu T, Sun H, et al. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and
degradation. Oncogene. 2018;37:5534–51. Article CAS PubMed Google Scholar * Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–58.
Article CAS PubMed Central PubMed Google Scholar * Seyrek K, Wohlfromm F, Espe J, Lavrik IN. The cross-talk of autophagy and apoptosis in breast carcinoma: implications for novel
therapies. Biochem J. 2022;479:1581–608. Article CAS PubMed Google Scholar * Tang Q, Su Z, Gu W, Rustgi AK. Mutant p53 on the path to metastasis. Trends Cancer. 2020;6:62–73. Article
PubMed Google Scholar * Castelli M, Piobbico D, Chiacchiaretta M, Brunacci C, Pieroni S, Bartoli D, et al. HOPS/TMUB1 retains p53 in the cytoplasm and sustains p53-dependent mitochondrial
apoptosis. EMBO Rep. 2020;21:e48073. Article CAS PubMed Google Scholar * Gorelick-Ashkenazi A, Weiss R, Sapozhnikov L, Florentin A, Tarayrah-Ibraheim L, Dweik D, et al. Caspases maintain
tissue integrity by an apoptosis-independent inhibition of cell migration and invasion. Nat Commun. 2018;9:2806. Article PubMed Central PubMed Google Scholar * Zhao J, Ou B, Han D, Wang
P, Zong Y, Zhu C, et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer. 2017;16:70.
Article PubMed Central PubMed Google Scholar * Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. Article CAS
PubMed Google Scholar * Venkatesh D, O’Brien NA, Zandkarimi F, Tong DR, Stokes ME, Dunn DE, et al. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev.
2020;34:526–43. Article CAS PubMed Central PubMed Google Scholar * Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, et al. Dual inhibition of MDMX and MDM2
as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10:eaao3003. Article PubMed Central PubMed Google Scholar * Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, et al. Targeting
USP7-mediated deubiquitination of MDM2/MDMX-p53 pathway for cancer therapy: are we there yet. Front Cell Dev Biol. 2020;8:233. Article PubMed Central PubMed Google Scholar * Munisamy M,
Mukherjee N, Thomas L, Pham AT, Shakeri A, Zhao Y, et al. Therapeutic opportunities in cancer therapy: targeting the p53-MDM2/MDMX interactions. Am J Cancer Res. 2021;11:5762–81. CAS PubMed
Central PubMed Google Scholar * Wang X, Wang J, Tsui YM, Shi C, Wang Y, Zhang X, et al. RALYL increases hepatocellular carcinoma stemness by sustaining the mRNA stability of TGF-β2. Nat
Commun. 2021;12:1518. Article CAS PubMed Central PubMed Google Scholar * Zhang RY, Liu ZK, Wei D, Yong YL, Lin P, Li H, et al. UBE2S interacting with TRIM28 in the nucleus accelerates
cell cycle by ubiquitination of p27 to promote hepatocellular carcinoma development. Signal Transduct Target Ther. 2021;6:64. Article CAS PubMed Central PubMed Google Scholar * Montal
R, Sia D, Montironi C, Leow WQ, Esteban-Fabró R, Pinyol R, et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol. 2020;73:315–27. Article CAS
PubMed Central PubMed Google Scholar * Wang D, Tian J, Yan Z, Yuan Q, Wu D, Liu X, et al. Mitochondrial fragmentation is crucial for c-Myc-driven hepatoblastoma-like liver tumors. Mol
Ther. 2022;30:1645–60. Article CAS PubMed Central PubMed Google Scholar * Beck A, Trippel F, Wagner A, Joppien S, Felle M, Vokuhl C, et al. Overexpression of UHRF1 promotes silencing of
tumor suppressor genes and predicts outcome in hepatoblastoma. Clin Epigenetics. 2018;10:27. Article PubMed Central PubMed Google Scholar * Makino Y, Hikita H, Fukumoto K, Sung JH,
Sakano Y, Murai K, et al. Constitutive activation of the tumor suppressor p53 in hepatocytes paradoxically promotes non-cell autonomous liver carcinogenesis. Cancer Res. 2022;82:2860–73.
Article CAS PubMed Central PubMed Google Scholar * Chen L, Liu S, Tao Y. Regulating tumor suppressor genes: post-translational modifications. Signal Transduct Target Ther. 2020;5:90.
Article CAS PubMed Central PubMed Google Scholar * Dinarvand N, Khanahmad H, Hakimian SM, Sheikhi A, Rashidi B, Bakhtiari H, et al. Expression and clinicopathological significance of
lipin-1 in human breast cancer and its association with p53 tumor suppressor gene. J Cell Physiol. 2020;235:5835–46. Article CAS PubMed Google Scholar * Chung SK, Zhu S, Xu Y, Fu X.
Functional analysis of the acetylation of human p53 in DNA damage responses. Protein Cell. 2014;5:544–51. Article CAS PubMed Central PubMed Google Scholar * Luo J, Li M, Tang Y,
Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101:2259–64. Article CAS PubMed Central
PubMed Google Scholar * Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001;2:1029–34. Article
CAS PubMed Central PubMed Google Scholar * Yang J, Jin A, Han J, Chen X, Zheng J, Zhang Y. MDMX recruits UbcH5c to facilitate MDM2 E3 ligase activity and subsequent p53 degradation in
vivo. Cancer Res. 2021;81:898–909. Article CAS PubMed Google Scholar * Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion
of p53. Nature. 1995;378:203–6. Article CAS PubMed Google Scholar * Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G. Mdm2 and Mdm4 loss regulates distinct p53 activities. Mol
Cancer Res. 2008;6:947–54. Article CAS PubMed Central PubMed Google Scholar * Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev
Cancer. 2014;14:359–70. Article CAS PubMed Central PubMed Google Scholar * Nagasaka M, Miyajima C, Aoki H, Aoyama M, Morishita D, Inoue Y, et al. Insights into regulators of p53
acetylation. Cells. 2022;11:3825. Article CAS PubMed Central PubMed Google Scholar * Xia Z, Kon N, Gu AP, Tavana O, Gu W. Deciphering the acetylation code of p53 in transcription
regulation and tumor suppression. Oncogene. 2022;41:3039–50. Article CAS PubMed Central PubMed Google Scholar * Gladka MM, Kohela A, Molenaar B, Versteeg D, Kooijman L,
Monshouwer-Kloots J, et al. Cardiomyocytes stimulate angiogenesis after ischemic injury in a ZEB2-dependent manner. Nat Commun. 2021;12:84. Article CAS PubMed Central PubMed Google
Scholar * Zeng F, Huang L, Cheng X, Yang X, Li T, Feng G, et al. Overexpression of LASS2 inhibits proliferation and causes G0/G1 cell cycle arrest in papillary thyroid cancer. Cancer Cell
Int. 2018;18:151. Article PubMed Central PubMed Google Scholar * Yang Y, Yang X, Lin Y, Yang G, Li L. LASS2 regulates hepatocyte steatosis by interacting with NDUFS2/OXPHOS related
proteins. Biochem Biophys Res Commun. 2020;526:871–9. Article CAS PubMed Google Scholar Download references FUNDING Funding This work was supported by the National Natural Science
Foundation of China (NSFC 81960494; NSFC 82002824; NSFC 82160484), Guizhou Province Science Plan Program (Qian Ke He Foundation ZK [2022] General 640), Science and Technology Fund Project of
Guizhou Provincial Health Commission (gzwjkj2019-1-192). AUTHOR INFORMATION Author notes * These authors contributed equally: Qingqing Zhao, Wei He. AUTHORS AND AFFILIATIONS * Department of
General Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, China Qingqing Zhao & Rui Chen * Department of Laboratory Medicine, Affiliated Hospital of Zunyi
Medical University, Zunyi, Guizhou, China Wei He, Zhouheng Liu, Xiaoli Yang, Xun Min & Yan Yang * School of Laboratory Medicine, Zunyi Medical University, Zunyi, Guizhou, China Wei He,
Zhouheng Liu, Xiaoli Yang, Xun Min & Yan Yang * Department of General Surgery, Shanghai Pudong Hospital, Fudan University Pudong Medical Center, Pudong, Shanghai, China Liangliang Huang
* School of Forensic Medicine, Zunyi Medical University, Zunyi, Guizhou, China Yong Liu & Yan Yang * Center of Forensic Expertise, Affiliated hospital of Zunyi Medical University, Zunyi,
Guizhou, China Yong Liu Authors * Qingqing Zhao View author publications You can also search for this author inPubMed Google Scholar * Wei He View author publications You can also search
for this author inPubMed Google Scholar * Zhouheng Liu View author publications You can also search for this author inPubMed Google Scholar * Liangliang Huang View author publications You
can also search for this author inPubMed Google Scholar * Xiaoli Yang View author publications You can also search for this author inPubMed Google Scholar * Yong Liu View author publications
You can also search for this author inPubMed Google Scholar * Rui Chen View author publications You can also search for this author inPubMed Google Scholar * Xun Min View author
publications You can also search for this author inPubMed Google Scholar * Yan Yang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS YY and
XM designed the study. Q-QZ and WH wrote the manuscript. YY, Q-QZ, Z-HL, WH, and L-LH performed the experiments. X-LY and RC performed data analysis. YL was responsible for the preparation
of pathology slides and analysis of immunohistochemistry results. XM provided valuable discussion and intellectual input. All authors provided critical feedback on manuscript and approved
the final version of the manuscript. CORRESPONDING AUTHORS Correspondence to Xun Min or Yan Yang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests.
ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION
FIGURE LEGENEDS FOR SUPPLEMENTARY FIGURE. S1 FIGURE. S2 FIGURE. S3 SUPPLEMENTAL TABLE 1-5 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0
International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and
the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative
Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by
statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zhao, Q., He, W., Liu, Z. _et al._ LASS2 enhances p53 protein stability and nuclear
import to suppress liver cancer progression through interaction with MDM2/MDMX. _Cell Death Discov._ 9, 414 (2023). https://doi.org/10.1038/s41420-023-01709-2 Download citation * Received:
25 July 2023 * Revised: 23 October 2023 * Accepted: 02 November 2023 * Published: 14 November 2023 * DOI: https://doi.org/10.1038/s41420-023-01709-2 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative