Hsp70 and hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Molecular chaperones such as Hsp40 and Hsp70 hold the androgen receptor (AR) in an inactive conformation. They are released in the presence of androgens, enabling transactivation
and causing the receptor to become aggregation-prone. Here we show that these molecular chaperones recognize a region of the AR N-terminal domain (NTD), including a FQNLF motif, that
interacts with the AR ligand-binding domain (LBD) upon activation. This suggests that competition between molecular chaperones and the LBD for the FQNLF motif regulates AR activation. We
also show that, while the free NTD oligomerizes, binding to Hsp70 increases its solubility. Stabilizing the NTD-Hsp70 interaction with small molecules reduces AR aggregation and promotes its
degradation in cellular and mouse models of the neuromuscular disorder spinal bulbar muscular atrophy. These results help resolve the mechanisms by which molecular chaperones regulate the
balance between AR aggregation, activation and quality control. SIMILAR CONTENT BEING VIEWED BY OTHERS LARGE-SCALE, IN-CELL PHOTOCROSSLINKING AT SINGLE-RESIDUE RESOLUTION REVEALS THE
MOLECULAR BASIS FOR GLUCOCORTICOID RECEPTOR REGULATION BY IMMUNOPHILINS Article 09 November 2023 CRYO-EM REVEALS HOW HSP90 AND FKBP IMMUNOPHILINS CO-REGULATE THE GLUCOCORTICOID RECEPTOR
Article Open access 09 November 2023 GLUCOCORTICOID RECEPTOR COMPLEXES FORM COOPERATIVELY WITH THE HSP90 CO-CHAPERONES PP5 AND FKBPS Article Open access 01 July 2020 INTRODUCTION Molecular
chaperones are key components of the protein quality control system. In addition to their well-known roles in protein folding and degradation1,2, they also stabilize specific conformations
of proteins to enable the fast and efficient relay of signaling information. This aspect of their function is exemplified by their activity on steroid hormone receptors (SHRs), such as the
glucocorticoid receptor (GR) and the androgen receptor (AR). It is known that a complex of heat shock proteins, including Hsp90, Hsp70, HOP, and Hsp40, is required to stabilize an inactive
SHR conformation3,4. This activity is termed a holdase function because the apo-receptor is held in a structural state that is stable, soluble and prepared for binding to ligands5. Hormone
binding then shifts the SHR into a distinct conformer of lower solubility that gains affinity for DNA and is capable of activating transcription. Intriguingly, at least some of the members
of the chaperone machinery become more transiently associated with the SHR during this process6, suggesting that these complexes are dynamic and play a role in stabilizing the conformation
of the active receptor. In addition, a subset of the chaperones, including Hsp70, is also required for SHR turnover, through recruitment of E3 ubiquitin ligases7. Thus, the SHR system
provides an excellent model for understanding how molecular chaperones balance the needs of protein folding, stability, function and quality control. Pioneering studies by Pratt et al. have
revealed the order of chaperone binding and release from SHRs3,4 and defined the importance of their individual functions in cell-free systems and in cells8. In support of this notion, a
recent study showed that Hsp70, HOP, and Hsp90 bind to the ligand-binding domain (LBD) of GR9, holding it in a stable, open conformation. However, it is unknown whether this mechanism is
conserved in other nuclear receptors; indeed, the Buchner group recently found that different Hsp90 co-chaperones are required for activating GR when compared to AR10. In addition the
structural roles of the chaperones during SHR activation and turnover are also not clear: for example, it has been proposed that Hsp70 interactions with the LBD are involved in receptor
triage11,12, but chemical modulators of Hsp70 or Hsp40 induce ubiquitin-dependent degradation of AR variants that lack the LBD13. These observations suggest that some chaperones, especially
Hsp70 and Hsp40, may have additional binding sites outside the canonical LBD region and that these might be critical for homeostasis of some SHRs. A mechanistic knowledge of how SHRs are
regulated by chaperones is also important for better understanding their roles in disease. For example, AR is an important therapeutic target for prostate cancer, because it controls the
expression of genes associated with proliferation14,15 in response to androgens, such as dihydrotestosterone (DHT)16. AR is also clinically important in spinal and bulbar muscular atrophy
(SBMA), a progressive degenerative disorder of the neuromuscular system caused by the pathologic expansion of a polyglutamine (polyQ) tract in the disordered, N-terminal domain of AR17. In
vitro and in cells, polyQ repeats beyond 37 residues in length cause hormone-dependent AR misfolding and aggregation. In patients and animal models, this misfolding manifests through the
formation of nuclear inclusions in motor neurons and skeletal muscle cells that contain ubiquitinated and misfolded polyQ AR18,19. Importantly, the chaperone-bound form of polyQ AR appears
to be protected from aggregation, such that it only becomes insoluble after androgen-mediated release of the chaperones. Together, these findings in prostate cancer and SBMA have created
particular interest in understanding the molecular mechanisms of how chaperones control the solubility, activation and turnover of AR8,12,20,21. Such discoveries could lead to new
therapeutics: for example, it has recently been shown that chemical activators of Hsp70 regulate the transcriptional activity of AR22,23. AR is a 919-residue protein composed of three
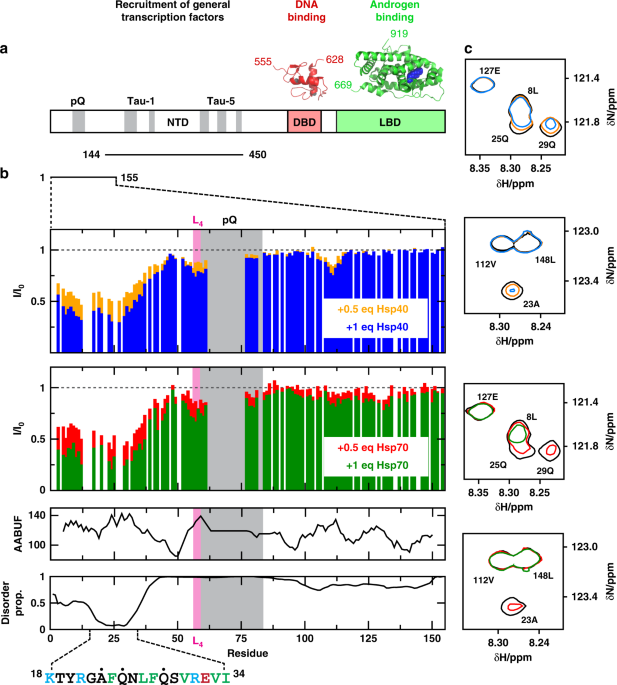
domains: the N-terminal transactivation domain (NTD; residues 1–558), the central DNA-binding domain (DBD; 559-619) and the C-terminal LBD (669–919). The structures of the DBD and LBD bound
to their ligands have been characterized by X-ray crystallography24,25,26,27. The NTD is intrinsically disordered28, but solution nuclear magnetic resonance (NMR) spectroscopy has shown that
it is rich in secondary structure, especially in regions key for transactivation29,30. The NTD of AR is one of the longest in the human SHR family and it includes the polyQ tract, starting
at residue 58, which becomes expanded in SBMA17. Binding of androgens to the LBD causes a large conformational change that leads to the nuclear localization of the receptor, driven by
exposure of a nuclear localization signal between the DBD and the LBD16. Despite its relevance for both transcriptional activity and the onset of SBMA, the nature of this conformational
change is not yet well-established. Nonetheless, it is known that androgen binding exposes a hydrophobic patch in the LBD, termed activation function 2 (AF-2), that has high affinity for a
5-residue FQNLF motif in the N-terminal segment of the NTD31. The inter-domain interaction between this highly conserved motif and AF-2 has been characterized by various biophysical methods,
including X-ray crystallography. These studies show that FQNLF binds to the LBD as an α-helix32, but it is not clear why this conformational transition is also associated with the
disassembly of the chaperone complex. It has been proposed that, similar to GR9, molecular chaperones bind to the LBD of AR and that their affinities for it decrease upon interaction with
DHT, but the mechanisms underlying this regulation are unknown. To investigate the earliest stages of AR activation by androgens, we analyzed the interaction of Hsp70 and Hsp40 with the NTD
of AR in vitro, by solution NMR33, and in cells. Solution NMR is particularly well-suited to the characterization of transient protein–protein interactions (PPIs) involving disordered
domains34. We found that Hsp70 and Hsp40 bind the NTD, with µM affinity and, strikingly, that the binding site contains the same FQNLF motif that interacts with AF-2 in the LBD during
activation. This observation suggests a tug-of-war model in which the FQNLF motif can be bound by either chaperone or the LBD, such that hormone binding shifts partitioning between these two
possibilities. In addition, we found that this part of the NTD has the propensity to form oligomers that are stabilized by interactions involving residues of the FQNLF motif, leading to
aggregation; however its interaction with Hsp70 keeps the protein in solution, explaining why chaperones are so well positioned to block the aggregation and misfolding of AR. Given that
Hsp40, Hsp70, and other chaperones bind near polyQ repeats in other proteins such as huntingtin this suggests a general mechanism35,36 by which these molecular chaperones keep proteins
harboring polyQ tracts soluble. Inspired by these findings, we used small molecules to stabilize the interactions of Hsp70 with AR containing an expanded polyQ tract in cellular and mouse
models of SBMA. Consistent with the results of the biophysical studies, these treatments reduce aggregation and enhance clearance of polyQ-expanded AR. Our results thus provide a structural
basis for how chaperones manage the stability, activation, solubility and turnover of AR, which should inform efforts to treat both prostate cancer and SBMA. RESULTS HSP70 AND HSP40 BIND TO
THE FQNLF MOTIF IN THE AR NTD We used solution NMR to determine whether Hsp40 and Hsp70 bind to the NTD of AR and, if so, where this interaction occurs. This study took advantage of recent
reports that characterized two NTD constructs (NTD1-155 and NTD144-450) in solution26,29. Briefly, NTD1-155 contains the FQNLF motif and the polymorphic polyQ repeat associated with SBMA,
and spans residues 1–155 according to numbering used in the AR entry in the Uniprot database (http://www.uniprot.org/uniprot/P10275); the version of NTD1-155 that we used contains 25
residues in the polyQ repeat, unlike the sequence found in Uniprot, that contains 23 (Fig. 1a). NTD144-450 contains the regions of the NTD involved in interactions with transcription factors
and transcriptional co-regulators, termed activation function 1 (AF-1)29,30. We incubated NTD1-155 (33 µM) or NTD144-450 (33 µM) with either 0.5 or 1 molar equivalents of human Hsp70
(HSPA1A) or Hsp40 (DNAJA2). These experiments were performed in the absence of added nucleotides and samples were held at 5 °C, a temperature that maximizes spectral quality. The 1H,15N-HSQC
spectrum of NTD144-450 was not altered by the addition of either Hsp40 or Hsp70, indicating weak or no interaction (Supplementary Fig. 1). This result is important because NTD144-450
includes hydrophobic motifs that could represent non-specific sites for chaperone binding, yet none were observed to interact under these conditions. By contrast, some signals in the
spectrum of NTD1-155 decreased in intensity (I/I0 ~ 0.3 at [Hsp]/[NTD1-155] = 1) without changing their position in the spectra (Fig. 1b, c). This result indicates that the chaperones
interact selectively with a region within NTD1-155. We found that the states populated under these conditions - the free state and, likely, various bound states37 - are in
intermediate-to-slow exchange on the NMR timescale. The low temperature used to carry out these experiments (5 °C) might contribute to the low observed exchange rate. Also this scenario is
typical for chaperone binding38,39 and suggests a dissociation constant lower than the concentration used in the experiments i.e. in the low micro molar range (see below). In these
experiments, clear decreases in signal intensities were observed in the first 35 residues of NTD1-155, which, strikingly, includes the motif FQNLF; this motif forms a complex with the LBD
after activation by androgens31,32. We observed similar changes for Hsp70 and Hsp40, which is consistent with the known similarity of consensus sites for these two chaperones (see
below)40,41. In addition, we observed much smaller decreases in signal intensity in other residues of NTD1-155 likely caused by transient interactions. For example, intensity changes were
observed in the hydrophobic 55LLLL58 motif that induces a helical structure in the polyQ tract of AR26,27. In summary we found that, prior to activation by androgens, Hsp70 and Hsp40 bind
to the NTD of AR by selectively interacting with a short region that includes the FQNLF motif. To determine whether this motif is an important part of the region that binds to Hsp40 and
Hsp70, we produced a mutant NTD1-155, named AQNAA-NTD1-155, in which residues Phe 24, Leu 27 and Phe 28 were mutated to Ala. These mutations were predicted to decrease the hydrophobicity of
the putative binding site, expressed as AABUF42 (Supplementary Fig. 2A), a property that is key for the ability of these molecular chaperones to recognize their substrates37,43. By NMR,
neither Hsp40 nor Hsp70 interacted with AQNAA-NTD1-155, confirming that the FQNLF motif is indeed important for the interaction between these molecular chaperones and this region of AR
(Supplementary Fig. 2A). There are two possible regions of Hsp70 where substrates bind to: a canonical site in a hydrophobic groove in the substrate binding domain (SBD) and a non-canonical
interaction surface44. To test which site was used by AR’s NTD1-155 we titrated it into solutions of Hsp70SBD (residues 394 to 509) and measured competition for a model, canonical peptide
(LVEALY-FAM) by fluorescence polarization (FP) in the absence of nucleotides. We found that NTD1-155 competed with LVEALY-FAM (Fig. 2), even better than the NR peptide, a bona fide Hsp70
substrate, suggesting that it bound with an IC50 1.7 ± 0.5 µM to the canonical binding site. We repeated these experiments by using the AQNAA-NTD1-155 mutant and found, in agreement with the
NMR experiments, that it had ~10-fold weaker affinity for Hsp70 (Supplementary Fig. 3). To further verify this interaction by an independent method, we measured the affinity between
NTD1-155 and Hsp70 by isothermal titration calorimetry (ITC). Although it was challenging to quantify the affinity because of solubility limitations, the binding affinity was estimated to be
~5 µM (Fig. 2b). This value is in good agreement with the FP studies as well as with the NMR titrations because this dissociation constant should lead, in the slow exchange regime, to I/I0
~ 0.3 when the NTD1-155 (at 33 µM) is in the presence of an equimolar amount of molecular chaperone (see Fig. 1b). Together, these results suggest that the region of sequence corresponding
to the FQNLF motif of AR and its flanking regions represents a canonical substrate for Hsp70. As mentioned above, Hsp40 and Hsp70 often compete for the same sites on their client proteins.
To understand the relative affinities of these chaperones, we measured binding of purified, human Hsp40 (DnaJA2) for NTD1-155 by ITC. Hsp40 bound slightly tighter to NTD1-155, with a Kd of
3.3 ± 2.3 µM (Supplementary Fig. 3B). The stoichiometry of the complex was estimated to be ~2 Hsp40s per NTD1-155, (_n_ = 0.49 ± 0.27) consistent with the dimeric structure of this
chaperone. These results suggest that both Hsp70 and Hsp40 bind with low micromolar affinity to NTD1-155. HSP70 AND HSP40 BIND TO THE FQNLF MOTIF IN CELLS To validate in cells the
observations made in vitro HEK293T cells were transfected with full length AR tagged with the FLAG epitope (FLAG-AR) and its interaction with endogenous Hsp40 and Hsp70 was assessed by
proximity ligation assay (PLA). This immunofluorescence-based technique is particularly useful for detecting transient PPIs in cells where the putative complexes are of intermediate or low
stability and therefore unlikely to withstand the washing steps that are required for co-immunoprecipitation experiments45. Briefly, the method relies on the use of primary antibodies that
recognize the two proteins (e.g. AR and Hsp70), and on secondary antibodies that are conjugated to complementary oligonucleotides. When the proteins are in close proximity, these
oligonucleotides hybridize and are ligated to form a circular DNA template that can be amplified by DNA polymerase and detected by fluorescence probes. The PLA signals can therefore be
detected by fluorescence microscopy as red puncta in the PLA experiment. Using this approach, we observed a clear interaction between FLAG-AR and both Hsp40 and Hsp70 in the cytoplasm prior
to activation but found that DHT treatment induced an almost complete loss of the interaction (Fig. 3a, c) combined with nuclear translocation (Supplementary Fig. 4). These results agree
with the notion that activation of AR by DHT causes the dissociation of the cytosolic complexes that AR forms with molecular chaperones prior to nuclear translocation3,8. To test whether the
interaction detected by PLA occurs in cells via the region of sequence containing the FQNLF motif detected by NMR, we repeated these experiments using a version of FLAG-AR with the FQNLF
motif mutated to AQNAA (FLAG-AR-mut) and a deletion mutant, where the region of sequence spanning residues 21 to 35, which includes the FQNLF as well as adjacent similar VREVI motif, was
deleted (FLAG-AR-del). We observed that while FLAG-AR-mut could still interact with both Hsp40 and Hsp70 (Supplementary Fig. 5), the interaction of FLAG-AR-del with both molecular chaperones
was substantially weaker (Fig. 3a, c). These results suggest that mutation of the three residues at the core of the FQNLF motif is not sufficient to abolish the affinity for Hsp70 or Hsp40,
that instead requires the complete deletion of this motif and the ca. 10 residues immediately flanking it. This result is consistent with the 10-fold weakened affinity of the AQNAA mutant
for Hsp70 observed in the NMR and FP experiments (see Supplementary Fig. 3). It should be noted that our PLA experiments were designed to detect the interaction of molecular chaperones with
the NTD of AR and are therefore unlikely to report on whether they interact with the LBD. In addition, the fact that both FLAG-AR-mut and FLAG-AR-del readily translocate to the nucleus upon
activation suggests that these variants retain some early AR functions (Supplementary Fig. 4). Collectively, these results confirm that, in cells, prior to activation by androgens, the
molecular chaperones Hsp40 and Hsp70 bind to AR via a region of sequence including the motif FQNLF and the residues flanking it. Hsp70 and Hsp40 have both been implicated in turnover of AR.
To probe the role of their interaction with the FQNLF motif in this process, we compared the levels of FLAG-AR-mut and FLAG-AR-del with those of the wild type protein prior and following to
activation with DHT. We observed that mutation and, especially, deletion of the FQNLF motif caused an increase in basal AR levels in the absence of hormone (Fig. 3b, Supplementary Fig 5B).
Also, recent studies have shown that disrupting AR’s N/C interaction delays disease onset in a mouse model of SBMA46. These studies utilized mice that expressed a mutant form of the polyQ AR
where the Phe residue of the FQNLF motif was mutated to Ala. To determine whether this mutation had an effect on the interaction between the AR NTD and molecular chaperones, we carried out
experiments with the equivalent mutant of NTD1-155, named AQNLF-NTD1-155. An analysis of the interaction by NMR showed that the AQNLF mutant interacts in vitro with molecular chaperones with
an affinity that was indistinguishable from that of the WT (Supplementary Fig. 6). Thus, our results confirm that amelioration of the SBMA phenotype in mice expressing this mutant is likely
due to a weakening of the N/C interaction rather than an altered interaction with molecular chaperones. HSP70 INCREASES THE SOLUBILITY OF THE AR NTD Because Hsp70 and Hsp40 are released
from the NTD1-155 after addition of the androgen dihydrotestosterone (DHT), the hormone-dependent phenotype of SBMA and ligand-dependent aggregation of the polyQ AR strongly suggest a
protective role for these molecular chaperones in limiting AR aggregation47,48,49,50. To investigate this possibility by solution NMR, we analyzed the oligomerization properties of the
NTD1-155 construct by measuring 1H,15N-HSQC spectra at two different concentrations, 125 and 450 μM at 278 K. NMR can be used to monitor concentration-dependent oligomerization because the
monomer-to-oligomer exchange process contributes to NMR linewidths, such that the _R_2 15N relaxation rates are increased51. In NTD1-155 we first confirmed that the position of the backbone
amide chemical shifts did not change with concentration. Then, we measured the _R_2 15N relaxation rates and found that some residues had higher values at 450 μM, when compared to 125 μM
(Fig. 4a). A closer examination of these linewidth changes showed that the affected residues were clustered into regions of locally high AABUF values, suggesting that hydrophobicity could be
the main driving force of oligomerization. Indeed, the most affected region was localized to the first 50 residues of the construct, with a maximum in the FQNLF motif. This observation is
conceptually equivalent to that made by Wetzel and co-workers in huntingtin, in which it was shown that the early stages of aggregation are not driven by inter-molecular interactions
involving the polyQ tract, but rather by hydrophobic, coiled coil-like interactions in the sequence flanking it at the N-terminus52,53. Further, these results raise the intriguing
possibility that binding of molecular chaperones to this region of AR might directly suppress its oligomerization. We then carried out NMR experiments to directly study whether the
interaction of Hsp70 with the NTD1-155 construct influenced its solubility. For these experiments, we prepared samples of NTD1-155 at 125 μM and added human Hsp70 at 1 molar equivalent,
using the same conditions presented in Fig. 1, and measured the corresponding 1H,15N-HSQC spectra as a function of incubation time at 298 K. In the absence of chaperone, we observed a
progressive decay of signal intensity, so that after 6 days of incubation, ~25 % of the signal intensity was lost. This signal decay was likely caused by formation of aggregates whose size
precluded their detection by NMR, as evident by the turbidity of the samples over time. By contrast, the samples containing Hsp70 did not show any decrease of signal intensity, suggesting
that the complex remained soluble (Fig. 4b). These results suggest that molecular chaperones limit self-interactions between FQNLF motifs in different AR NTD molecules, diminishing its
oligomerization and maintaining solubility. HSP70 INCREASES AR SOLUBILITY IN VIVO A key prediction of these studies is that chemical stabilization of the Hsp70 interaction might limit
aggregation of polyQ AR. Recent work has produced chemical probes that significantly enhance Hsp70’s affinity for disordered proteins54. Briefly, Hsp70 normally cycles through rounds of ATP
hydrolysis, with the ATP-bound state having relatively poor affinity for substrate proteins and the ADP-bound state having tight affinity55. These compounds, such as YM-1, favor the
ADP-bound state56, therefore stabilizing Hsp70’s interactions. In addition, stabilizing this complex has been shown to enhance turnover of some Hsp70 client proteins54. Thus, we hypothesized
that these compounds might limit polyQ AR aggregation and potentially promote its turnover in SBMA models. To understand if Hsp70 modulators might have these favorable activities, we first
tested a small panel of them in PC12 cells that stably express AR with an expanded polyQ tract encoded by 112 CAG repeats, termed AR112Q, under a tetracycline responsive promoter57. In these
cells, treatment with ligands, such as DHT or the synthetic androgen metribolone (R1881), leads to nuclear accumulation of misfolded polyQ AR, consistent with aggregation after release of
chaperones. SDS-insoluble polyQ-AR species can be separated by centrifugation and visualized by western blot or by filter trap assay (Supplementary Fig. 7). In our small-scale screen, we
found that two Hsp70 modulators, JG-84 and JG-98, were the most potent in enhancing solubility of polyQ AR (Supplementary Fig. 7A-D). Moreover, the active molecules promoted clearance of
both monomeric and aggregated polyQ AR species (Supplementary Figs. 7D–F, 8), and were more striking on polyQ AR than wild type AR (Supplementary Fig. 8). Further analysis demonstrated that
the more potent Hsp70 modulator, JG-98, increased polyQ AR ubiquitination and enhanced degradation through the proteasome (Fig. 5a, b). Notably, treatment with JG-258, a structural analog of
JG-98 that does not bind Hsp7058, does not change steady state polyQ AR levels or alter polyQ AR ubiquitination (Supplementary Fig. 9). We wondered whether these effects might occur because
of an HSF1 dependent stress response. To test this idea, we performed western blots for three stress-inducible biomarkers: Hsp25, Hsp40, and Hsp70. In treated cells, reduction of AR112Q
occurred without induction of heat shock proteins (Fig. 5c). In contrast, the Hsp90 inhibitor, 17-AAG, strongly increased the levels of these proteins (Fig. 5c), consistent with induction of
an HSF1 dependent stress response. Additionally, JG-98 did not affect levels of Hsp90 client proteins in their native conformations, such as Akt and Erk (Fig. 5c), both of which are
degraded following treatment with Hsp90 inhibitors. Finally, recent work has suggested that combining Hsp70 modulators with Hsp90 inhibitors might further promote AR clearance12. Consistent
with this notion, we found that combinations of an Hsp70 modulator, JG-98, with an Hsp90 inhibitor, 17-AAG, produced robust polyQ AR clearance (Fig. 5d). Together, these findings supported a
model in which stabilizing the Hsp70-AR interaction enhances the solubility and clearance of AR. To test this idea in an animal model of SBMA, we turned to an analog of the Hsp70
modulators, JG-294, that is tolerated in mice and has well-studied pharmacokinetics58. Before proceeding into animals, we first confirmed that JG-294 could induce degradation of polyQ AR in
PC12 cells expressing AR112Q. Indeed, treatment with JG-294 yielded a significant reduction in the levels of AR112Q, as detected by western blot and immunofluorescence microscopy (Fig. 6b,
c). Next, we tested whether JG-294 might have activity in a well-characterized mouse model of SBMA generated by gene targeting (AR113Q mice)50,59. In this model, like the cell-based system,
AR113Q accumulates as SDS-insoluble high molecular weight species in skeletal muscle from mutant males. Strikingly, treatment with JG-294 for two weeks (i.p., at 3 mg kg−1 every other day)
reduced levels of both high molecular weight and monomeric polyQ AR when compared to the vehicle treated controls (Fig. 7a, b, d, e). These changes occurred without alteration in AR mRNA
levels (Fig. 7c), consistent with the notion that JG-294 promoted polyQ-AR clearance. Notably, as in cell culture, these effects were distinct from those triggered by treatment with an Hsp90
inhibitor, as no induction of Hsp70 or loss of client proteins in their native conformation were seen following treatment with JG-294 (Fig. 7e). Together, these results suggest that
stabilizing the interactions between Hsp70 and AR may be a viable therapeutic strategy for SBMA. Collectively, our findings support a model in which Hsp70 directly binds to the NTD of AR as
a sensor for misfolding and aggregation. DISCUSSION We show here that AR activation seems to be governed, in part, by a competition between chaperones and the AF-2 region for binding to a
region of the NTD including the FQNLF motif. Specifically, inter-molecular interactions with Hsp70 and Hsp40 appear to prevent activation of AR by sequestering the FQNLF motif while
intra-molecular interactions of this same region with AF-2 in the LBD are required for activation. We find that the relative strength of these competing PPIs is regulated by androgen binding
to the LBD, which stabilizes the AF-2, and, therefore, promotes its interaction with the FQNLF motif and displaces the chaperones. What is the role of the Hsp40 in this process? Hsp40 and
Hsp70 normally work in concert and, by analogy with other systems, we propose that Hsp40 binds AR first and then recruits Hsp70 via its conserved J-domain. In cells, binding of the J-domain
stimulates ATP cycling by Hsp7060, which creates the tight affinity, client-chaperone complex. Therefore, we expect that Hsp40 would assist Hsp70 by both localizing it to AR and accelerating
its catalytic cycle. This is important because our in vitro experimental platforms, while aimed at investigating the equilibrium properties of the system in the absence of nucleotide,
likely underestimate the effective affinity for the NTD. Moreover, there are more than 45 proteins in the Hsp40 family in humans, so the relevant subset for AR homeostasis is not yet clear.
One of the interesting implications of this model is that the structure of the FQNLF motif itself might switch when bound to either the chaperones or AF-2. Specifically, it is known that the
FQNLF motif is helical after binding to AF-2 and activation, yet we have shown that it can interact with the canonical groove of Hsp70, likely in an extended conformation. Thus, these
studies suggest that (at least) two sets of competing PPIs, in addition to a local conformational switch, underlie the activation of AR. Further experiments will be needed to understand
whether there is a link between these molecular events and the broader changes that occur during nuclear translocation and transcriptional activity of AR. Additionally, it has previously
been observed that Hsp70 binds to the LBD of SHRs9 and it is not yet known whether the N-terminal mechanism presented here is connected to other chaperone-mediated processes at the LBD.
Despite these remaining questions, this molecular view of AR dynamics and PPIs has a number of interesting implications. First, these results also provide insight into how Hsp70 and Hsp40
regulate the solubility of AR. We noted that Hsp70 prevents the oligomerization of AR’s NTD1-155 (Fig. 4), likely by directly encumbering the FNQLF motif and surrounding regions that are
hydrophobic and aggregation-prone. We speculate that this activity, combined with the fast turnover of AR prior to androgen binding, conspire to minimize the propensity of the receptor to
oligomerize and aggregate by preventing exposure of the most aggregation-prone sequence while simultaneously keeping its concentration low in the cytoplasm. In this scenario, molecular
chaperones would contribute, by acting as holdases, to keeping the receptor monomeric and at low concentration i.e. primed for rapid activation by androgens without compromising solubility.
Putting these mechanisms together, it seems elegant to propose that Hsp70’s interactions with the FQNLF motif might create a three-way tug-of-war, in which the chaperone competes for both:
(a) proper intra-molecular contacts with AF-2 and (b) inter-molecular contacts between the FQNLF motifs of two AR molecules, leading to aggregation. The relative affinities of these
competing PPIs likely balances the activation of the receptor and governs its unnatural aggregation in SBMA (Fig. 8). Based on the experiments with Hsp70 modulators, we propose that this
delicate balance of PPIs also regulates AR turnover. In other systems, such as microtubule-associated protein tau (MAPT) and X-linked inhibitor of apoptosis (XIAP), tight binding of Hsp70
has been observed to promote turnover54,61. Similarly, prolonged interactions with Hsp70 seem to favor turnover of polyQ-expanded AR. Thus, more broadly, Hsp70 seems to use a dwell time
method to discriminate between damaged and healthy proteins (Fig. 8). In the case of AR, the incorrect decision to release polyQ AR may partially underlie protein aggregation in SBMA, such
that chemical perturbations with JG-98 or JG-294, can partially re-balance the system and promote turnover. In this model, it is striking that the same chaperone that governs receptor
activation and solubility is also tasked with monitoring its quality control. Notably, numerous functions essential for cell viability, ranging from energy and ion homeostasis to axonal
transport and gene expression, are disrupted by misfolded proteins that cause age-dependent protein aggregation neurodegenerative disorders, including SBMA62,63. Alterations in these
multiple pathways likely contribute to toxicity in a cumulative manner, suggesting that therapeutically targeting any one of them is likely to yield incomplete rescue. In contrast,
decreasing levels of mutant proteins such as the polyQ AR has emerged as an attractive therapeutic strategy to target the proximal mediator of disease pathogenesis. In this regard, our
findings have implications for potential new therapies. Many efforts to develop chaperone-targeted therapies for protein aggregation diseases have focused on inhibition of Hsp90. Our data
indicate that targeting Hsp70 may have significant advantages. For example, Hsp70 activation favors the removal of misfolded proteins rather than client proteins in their native or
near-native states11,12,64. This distinction suggests that Hsp70-targeted small molecules may avoid some of the toxic effects of Hsp90 inhibitors, which promote degradation of hundreds of
client proteins in their native conformations, some of which might be needed for normal cellular functions. Similarly, the other major strategy for activating chaperone function is to target
the stress response transcription factor, HSF1. However, experimental evidence suggests that chronic activation of HSF1 may promote toxicity by triggering a maladaptive stress response that
is present in many protein-folding diseases65. In contrast, we found that targeting Hsp70 does not lead to activation of stress-responsive proteins. Thus, these results suggest that Hsp70
may be a good target for the treatment of SBMA. In addition, AR has been implicated in other diseases, such as prostate cancer. Indeed, recent studies have established that the same
Hsp70-targeted small molecules promote clearance of an AR splice variant that is associated with prostate cancer and lacks the C-terminal LBD13. Together, these observations point to Hsp70
as a promising target in multiple AR-associated diseases. Such strategies may take advantage of the normal protein quality control functions of this chaperone, which are already evolved to
regulate AR solubility and degrade the damaged receptor. METHODS PROTEIN EXPRESSION AND PURIFICATION The genes codifying for NTD1-155 and its variants were purchased from GeneArt and cloned
in a pDEST-HisMBP vector (Addgene: #11085) whereas that codifying for NTD144-45029 was instead cloned into a Gateway pDEST17 vector (Invitrogen). Expression of the resulting genes led to
fusion proteins containing a His6 tag, a maltose binding protein (MBP) moiety for NTD1-155 and its variants and a motif recognized by the tobacco etch virus (TEV) protease. The His6 tag was
used as a purification tag, whereas MBP was used to increase the solubility of NTD1-155 and its variants. 15N labeled protein expression was carried out in Rosetta (DE3) pLysS E. coli
competent cells Novagen (Merck), grown in Luria Broth (LB; Melford) at 37 °C at 220 r.p.m. until the value of OD600 was 0.7, centrifuged at 2000 G for 20 min, then washed in MOPS buffer
(3-(N-morpholino) propanesulfonic acid (200 mM), sodium acetate (50 mM), EDTA (10 mM), pH 7.0) devoid of nitrogen sources and rapidly centrifuged at 5000 G for 10 minutes. Cells were then
resuspended in MOPS media containing 15NH4Cl as a nitrogen source (Eurisotope), induced with 0.5 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG, Sigma) for 4 hours at 28 °C, harvested by
centrifugation and resuspended in core buffer (50 mM sodium phosphate, 500 mM NaCl, 5% (v/v) Glycerol, 1 mM 2-Mercapto-ethanol, pH 8.0). A HisTrap HP 5 ml column (GE Healthcare) was used to
purify the proteins, which were eluted by an imidazole gradient (final composition: 500 mM imidazole, 50 mM sodium phosphate, 500 mM NaCl, 5% Glycerol, 1 mM βMercapto-ethanol, pH 8.0),
followed by a size exclusion step carried out in a Superdex HighLoad S200 26/60 column (GE Healthcare) equilibrated in a buffer with the following composition: 500 mM NaCl, 12 mM sodium
phosphate, 5% glycerol, 1 mM DTT, pH 7.5. To separate NTD1-155 from its tags, the pure fusion proteins were then incubated with His6-tagged TEV protease for 16 hours at 4 °C by dialysis
against a buffer containing 20 mM sodium phosphate, 100 mM NaCl, and 0.5 mM EDTA, pH 8.0. The product of proteolytic cleavage was separated from the tags by Ni2+ affinity chromatography,
employing a buffer containing 8 M urea (500 mM imidazole, 50 mM sodium phosphate, 100 mM NaCl, 8 M urea, pH 8.0) to prevent the aggregation of the cleaved AR in the column. Finally, the
cleaved proteins were stored at −80 °C. Full length, human Hsp70 (HSPA1A) and Hsp40 (DnaJA2) constructs were transformed into BL21(DE3) cells and single colonies were used to inoculate
Terrific Broth (TB) medium containing ampicillin (50 µg mL−1). Cultures were grown at 37 °C for 5 h, cooled to 20 °C and induced overnight with 200 μM IPTG. DnaJA2 and Hsp70 (SBD) (residues
394–540) were purified using a Ni2+-NTA column, followed by overnight TEV cleavage of the His tag and gel filtration on a Superdex S200 (GE Healthcare). Hsp70 was purified using a Ni2+-NTA
column, followed by overnight TEV cleavage and subjection to an ATP agarose column. Hsp70 was eluted with ATP and dialyzed to remove excess nucleotide. This purification method provides
Hsp70 in its ADP-bound state, based on partial proteolysis. A truncated form of Hsp70SBD (HSPA1A) was used for the FP studies, which encompasses residues 394-509, was purified in a similar
way. NMR SAMPLE PREPARATION The protein solutions stored at −80 °C were thawed and dialyzed for 16 h at 4 °C against a phosphate buffer (sodium phosphate (20 mM),
tris(2-carboxyethyl)phosphine (TCEP, 1 mM), pH 7.4). Finally, 10% D2O and 0.015 mM DSS were added to the samples. ACQUISITION AND ANALYSIS OF NMR SPECTRA Two-dimensional 1H,15N-HSQC NMR
experiments were recorded at 5 °C on a Bruker 600 or 800 MHz spectrometer equipped with cryogenic probes. Spectra were processed with NMRPipe66 and analyzed using CcpNMR Analysis67. The NMR
samples used for the experiments reported in Fig. 1, Supplementary Figs. 1 and 5 contained 33 µM 15N single-labelled NTD1-155 protein and either 16.5 µM or 33 μM unlabeled Hsp70 and Hsp40 in
20 mM phosphate buffer, pH 7.4, 1 mM TCEP, and 10% (v/v) D2O. The NMR samples used for the experiments shown in Supplementary Fig. 1 were equivalent to those used for the experiments shown
in Fig. 1, but contained NTD144-450 instead of NTD1-155. The NMR samples used for the experiments reported in Fig. 4a contained 125 and 450 µM 15N single-labelled NTD1-155 constructs and
those used for the experiments reported in Fig. 4b contained 125 µM 15N single-labelled NTD1-155 constructs and 125 μM unlabeled Hsp70. The 15N _R_2 rates of NTD1-155 were measured with 10
different relaxation delays (t) and fit to the equation \(I = I_o{\mathrm{exp}}( - R_2t)\) and only the values of _R_2 determined with less than 5% error were considered sufficiently
reliable for the analysis shown in Fig. 4a. The ratio of signal intensities shown in Fig. 4b corresponds to the total signal intensity of the signals that are not affected by chaperone
binding (Fig. 1) measured at a given time divided by that measured after 1 day of equilibration. FLUORESCENCE POLARIZATION ASSAYS For competition FP studies, 2 μM Hsp70SBD and 25 nM
FAM-LVEALY (Anaspec) were incubated with varying concentrations of AR protein for 30 min at room temperature in assay buffer (100 mM Tris, 20 mM KCl, 6 mM MgCl2 pH 7.4). After incubation,
fluorescence polarization was measured (excitation: 485 nm emission: 535 nm) using a SpectraMax M5 plate reader. In the case of the FAM-NRLLLTG tracers, 12 μM Hsp70SDB, and 25 nM tracer were
used and experiment was performed in a similar manner. These experiments were repeated in duplicate and repeated on three independent days. ISOTHERMAL TITRATION CALORIMETRY The NTD1-155
proteins, as well as Hsp70SBD or Hsp40, were dialyzed overnight against ITC buffer (25 mM HEPES, 5 mM MgCl2, 10 mM KCl pH 7.5). Concentrations were determined using a BCA Assay (Thermo
Fisher Scientific), and the experiment was performed with a MicroCal ITC200 (GE Healthcare) at 25 °C. AR protein (100–200 μM) in the syringe was titrated into a 10–20 μM cell solution of
Hsp70 (SBD) protein. Calorimetric parameters were calculated using Origin 7.0 software and fit with a one or two-site binding model. Experiments were performed in duplicate and repeated
twice on two independent days. CELL CULTURE AND TRANSFECTION Human embryonic kidney 293 T cells (HEK293T cells) were obtained from ATCC (CRL-3216) and maintained in DMEM containing 4.5 g L−1
glucose (Glutamax, Gibco) supplemented with 10% (v/v) charcoal stripped calf serum (Gibco) and antibiotics. Cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C.
Transient transfection of HEK293T cells was performed with polyethylenimine (PEI) (Polysciences) at a ratio of 1 µg DNA to 3 µl PEI to express androgen receptor tagged with the FLAG epitope
(FLAG-AR), the mutated version (FLAG-mut) and the version lacking the region of sequence targeted by Hsp40 and Hsp70 in the NMR experiments (FLAG-del). PC12 cells expressing
tetracycline-inducible AR112Q (gift of Diane Merry, Thomas Jefferson University) were characterized previously57. Cells were maintained in phenol red-free DMEM (21063-029, Invitrogen)
supplemented with 5% charcoal stripped fetal calf serum (Thermo Scientific Hyclone Products), 10% charcoal stripped horse serum (Invitrogen), G418 (Gibco) and Hygromycin B (Invitrogen). When
indicated, cells were transfected using Lipofectamine 2000 (Life Technologies) or the Neon transfection system (Thermo Scientific) to express human CHIP and HA-His-ubiquitin (gifts from Dr.
Yoichi Osawa, Univ. of Michigan). AR expression was induced with 0.5 µg mL−1 doxycycline (Clonetech) and activated by treatment with the synthetic androgen R1881. Lactacystin was from
Sigma-Aldrich (L6785) and 17-AAG was from LC Laboratories (A-6880). ANTIBODIES AND REAGENTS For Western Blot analyses the following antibodies were used: anti-β-actin-HRP (ab8224, 1:10000)
(Abcam), rabbit-anti-FLAG (F7425, 1:2000), and mouse-anti-FLAG M2 (F1804, 1:2000) (Sigma), mouse-anti-HSP70/72 (ADI-SPA-810) and rabbit-anti-HSP40 (ADI-SPA-400-D) (Enzo Life Science).
Additional primary antibodies were used against Akt (C6E7; Cell Signaling Technology), ERK1/2 (9102, Signaling Technology), GAPDH (NB-600-502; Novus Biologicals), HSP90 (H114, sc7947; Santa
Cruz Biotechnology), CHIP (PA5-32046, Thermo Scientific, Pierce), HA (MMS-101R, Covance, Biolegend), HSP25 (ADI-SPA-801-488; Enzo Life Science), and HSP70 (ADI-SPA-812; Enzo Life Science).
HRP conjugated secondary antibodies were from Bio-Rad (170-6515, 170-6516). For immunofluorescence the following primary antibodies were used: rabbit-anti-FLAG (F7425, 1:200),
mouse-anti-FLAG M2 (F1804, 1:200) (Sigma), AlexaFluor conjugated secondary antibodies were from Life Technologies (A11072, 1:300). Mounting medium with DAPI (H-1200, 1:500) was from Vector
Labs. For immunoprecipitation, pull down was performed with protein A–agarose beads (Santa Cruz Biotechnology) and the anti-AR antibody (PG21; Millipore). WESTERN BLOTTING Cells were washed
and harvested in PBS, lysed in RIPA buffer (Technova) containing phosphatase and protease inhibitors (Roche). Lysates were centrifuged at 15000 g to separate soluble and pellet fractions.
Total protein was quantified using BCA assay (Pierce Biotechnology). Proteins were resolved by 7.5 or 15% SDS-PAGE, transferred to PVDF or nitrocellulose membranes, and probed with specific
antibodies. Filter retardation assay was performed by sample filtration through a 0.2 μm cellulose acetate membrane (Whatman, GE Healthcare) using a slot-blot apparatus. For ubiquitination
studies, cells were homogenized in buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol, pH 7.4) containing protease inhibitors and 5 mM N-ethylmaleimide (Sigma-Aldrich).
Lysates were incubated on a rotator at 4 °C for 2 h with AR antibody or non-immune rabbit IgG (Santa Cruz Biotechnology). Prewashed protein A–agarose beads were added, and samples were
incubated for 1 additional hour at 4 °C. Protein-antibody-bead complexes were washed 6 times in buffer, and proteins were eluted by boiling in SDS-loading buffer for 5 min at 100 °C. Samples
were then resolved on by 7.5% SDS-PAGE. Images of uncropped and unprocessed scans of blots are provided as Source Data files. Agarose gel electrophoresis for resolving aggregates was
performed as previously described68. Briefly 150 µg total protein was loaded to 1.5% agarose gels (375 mM Tris buffer, pH 8.6; 0.1% SDS) and electrophoresed at 100 V until the dye front had
reached the bottom of the gel. Transfer of proteins was performed using a semi-dry transfer apparatus to PVDF membranes. IMMUNOFLUORESCENCE For immunofluorescence, cells were seeded on
sterilized 12 mm microscope coverslips, fixed 15 min in 4% paraformaldehyde at room temperature and permeabilized in ice-cold methanol for 10 min. Cells were then washed, blocked with PBS
containing 2% bovine serum albumin for one hour at room temperature and incubated with the corresponding primary antibody diluted in blocking solution for one hour. After washing with PBS,
samples were further incubated with the corresponding secondary antibodies for one hour. DAPI was used to label DNA and the coverslips were washed, air-dried and mounted on slides using
Prolong Gold antifade mountant (Life technologies). 6 μm cryosections of muscle were fixed in methanol. Slides were stained, mounted, and imaged by confocal microscopy (Olympus FV 500 or
Nikon A1). Nuclear signal intensity was quantified by CellProfiler. PROXIMITY LIGATION ASSAY The Duolink in situ Proximity ligation (PLA) kit (Olink Bioscience) was used for the detection
of protein–protein interactions according to the manufacturer’s instructions45. The assay is based on the simultaneous binding of two antibodies to the proteins of interest and makes use of
a proximity ligation technique combined with an amplification reaction to detect the antibodies whenever they are less than 30 nm apart. HEK293T cells were grown and transfected as indicated
on 12 mm microscope coverslips (Thermo). Cells were fixed in PBS containing 4% paraformaldehyde for 15 min and permeabilized with methanol for 5 min. Fixed cells were blocked in PBS
containing 2% BSA (Sigma) for an hour and then incubated with the indicated antibodies against Hsp70 (ADI-SPA-810, 1:100), Hsp40 (ADI-SPA-400-D, 1:100) (Enzo Life Science) and FLAG (M2,
F1804 and F7425, both 1:50) (Sigma). Samples were subsequently incubated for 1 h at 37 °C with secondary antibody PLA with and without probes for mouse and rabbit (Sigma), and then for 30
min at 37 °C with a ligation solution to ligate the two PLA probes and form a circular DNA template. A rolling-circle amplification was then performed by incubating the cells for 100 min at
37 °C with the amplification solution. The amplicons generated are recognized by fluorescent oligonucleotides present in the amplification solution. Nuclei were stained with DAPI and the
coverslips were washed, air-dried and mounted on slides using Prolong antifade mountant (Life Technologies). Cells were analyzed with a ×63 objective on a Leica SP5 confocal microscope.
Images were presented as maximal projection of z-stacks by using Fiji. AR113Q MICE AR113Q knock-in mice were generated using exon 1-specific gene targeting50,59, backcrossed to C56BL/6 (≥10
generations), and housed in a specific pathogen-free facility on a 12-h light/dark cycle with chow and water ad libitum. For analysis, AR113Q males were used at 3 months of age and randomly
assigned to treatment groups. JG-294 was dissolved in 70% propylene glycol (W294004, Sigma-Aldrich) and 30% PBS (pH 7.4, Life Technologies), heated to 95 °C to dissolve compound, aliquoted
and frozen. Mice received intraperitoneal injections (3 mg kg−1) every other day for 2 weeks. 17-DMAG (S1142, Selleckchem) was dissolved in 1% DMSO (D2650, Sigma-Aldrich), 30% polyethylene
glycol (202371; Sigma-Aldrich) and 1% Tween80 (P4780; Sigma-Aldrich), and administered by i.p. injection (10 mg kg−1) every other day for 2 weeks. At the end of treatment, mice were
euthanized and quadriceps were harvested and flash frozen in liquid nitrogen or mounted on OCT (Tissue-Plus, Fisher Health Care) for cryosectioning. For western blot, lysates were obtained
by homogenizing tissue in RIPA containing phosphatase and protease inhibitors. All procedures involving mice were approved by the University of Michigan Committee on Use and Care of Animals
(PRO00008133) and conducted in accordance with institutional and federal ethical guidelines for animal testing and research. QUANTITATIVE RT-PCR Total RNA was extracted from mouse quadriceps
muscles using Trizol (Sigma-Aldrich). cDNA was synthesized from extracted RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Bio Systems). Quantitative PCR was carried out
on a 7500 Real-Time PCR System (Applied Biosystems) using FastStart TaqMan Probe Master Mix (Roche) and gene specific primers for AR (universal probe library #58, Roche; 5′ primer:
ccagtcccaattgtgtcaaa; 3′ primer: tccctggtactgtccaaacg) and Cpsf2 (Mm00489754_m1, Applied Biosystems). Relative AR expression was determined by the standard curve method and normalized to
Cpsf2. STATISTICS Statistical significance was determined by analyzing data sets using unpaired Student’s _t-_test, one-way ANOVA with Tukey’s multiple comparison test or two-way ANOVA with
Bonferroni or Niewman–Keuls multiple comparison test in Prism6 (GraphPad). Differences between mean values were defined as significant at _p_ < 0.05. REPORTING SUMMARY Further information
on research design is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The source data underlying the quantifications shown in Figs. 2A, 3B, 4B,
5A, 5D, 6B, 6C, 7B, 7C, 7E, and Supplementary Figs. 3A, 7F, and 9A are available as Source Data files and any other data is available from the corresponding authors upon reasonable request.
CHANGE HISTORY * _ 07 OCTOBER 2019 An amendment to this paper has been published and can be accessed via a link at the top of the paper. _ REFERENCES * Saibil, H. Chaperone machines for
protein folding, unfolding and disaggregation. _Nat. Rev. Mol. Cell Biol._ 14, 630–642 (2013). Article CAS Google Scholar * Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M. &
Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. _Annu. Rev. Biochem._ 82, 323–355 (2013). Article CAS Google Scholar * Pratt, W. B. & Toft, D. O.
Steroid receptor interactions with heat shock protein and immunophilin chaperones. _Endocr. Rev._ 18, 306–360 (1997). CAS PubMed Google Scholar * Pratt, W. B. & Toft, D. O. Regulation
of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. _Exp. Biol. Med._ 228, 111–133 (2003). Article CAS Google Scholar * Bohen, S. P., Kralli, A.
& Yamamoto, K. R. Hold’em and fold’em: chaperones and signal transduction. _Science_ 268, 1303 (1995). Article CAS ADS Google Scholar * Cato, L. et al. Development of Bag-1L as a
therapeutic target in androgen receptor-dependent prostate cancer. _eLife_ 6, e27159 (2017). * He, B. et al. An androgen receptor NH2-terminal conserved motif interacts with the COOH
terminus of the Hsp70-interacting protein (CHIP). _J. Biol. Chem._ 279, 30643–30653 (2004). Article CAS Google Scholar * Cano, L. Q., Lavery, D. N. & Bevan, C. L. Mini-review:
foldosome regulation of androgen receptor action in prostate cancer. _Mol. Cell. Endocrinol._ 369, 52–62 (2013). Article Google Scholar * Kirschke, E., Goswami, D., Southworth, D.,
Griffin, P. R. & Agard, D. A. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. _Cell_ 157, 1685–1697 (2014). Article CAS Google
Scholar * Sahasrabudhe, P., Rohrberg, J., Biebl, M. M., Rutz, D. A. & Buchner, J. The plasticity of the Hsp90 co-chaperone system. _Mol. Cell_ 67, 947–961.e5 (2017). Article CAS
Google Scholar * Pratt, W. B., Gestwicki, J. E., Osawa, Y. & Lieberman, A. P. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative
diseases. _Annu. Rev. Pharmacol. Toxicol._ 55, 353–371 (2015). Article CAS Google Scholar * Wang, A. M. et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein
degradation. _Nat. Chem. Biol._ 9, 112–118 (2013). Article CAS Google Scholar * Moses, M. A. et al. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat
castration-resistant prostate cancer. _Cancer Res._ 78, 4022–4035 (2018). Article CAS Google Scholar * Heinlein, C. A. & Chang, C. Androgen receptor in prostate cancer. _Endocr. Rev._
25, 276–308 (2004). Article CAS Google Scholar * Quigley, D. A. et al. Genomic hallmarks and structural variation in metastatic prostate cancer. _Cell_ 174, 758–769.e9 (2018). Article
CAS Google Scholar * Gelmann, E. P. Molecular biology of the androgen receptor. _J. Clin. Oncol._ 20, 3001–3015 (2002). Article CAS Google Scholar * Spada, A. R. L., Wilson, E. M.,
Lubahn, D. B., Harding, A. E. & Fischbeck, K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. _Nature_ 352, 77–79 (1991). Article ADS Google Scholar
* Li, M. et al. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. _Ann. Neurol._ 44, 249–254 (1998). Article CAS Google Scholar * Stenoien, D.
L. et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. _Hum. Mol.
Genet._ 8, 731–741 (1999). Article CAS Google Scholar * Prescott, J. & Coetzee, G. A. Molecular chaperones throughout the life cycle of the androgen receptor. _Cancer Lett._ 231,
12–19 (2006). Article CAS Google Scholar * Azad, A. A., Zoubeidi, A., Gleave, M. E. & Chi, K. N. Targeting heat shock proteins in metastatic castration-resistant prostate cancer.
_Nat. Rev. Urol._ 12, 26–36 (2014). Article Google Scholar * Dong, J. et al. Hsp70 binds to the androgen receptor N-terminal domain and modulates the receptor function in prostate cancer
cells. _Mol. Cancer Ther._ 18, 39–50 (2019). Article CAS Google Scholar * Liu, C. et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in
advanced prostate cancer. _Nat. Commun._ 9, 4700 (2018). Article ADS Google Scholar * Matias, P. M. et al. Structural evidence for ligand specificity in the binding domain of the human
androgen receptor: implications for pathogenic gene mutations. _J. Biol. Chem._ 275, 26164–26171 (2000). Article CAS Google Scholar * Shaffer, P. L., Jivan, A., Dollins, D. E., Claessens,
F. & Gewirth, D. T. Structural basis of androgen receptor binding to selective androgen response elements. _Proc. Natl Acad. Sci. USA_ 101, 4758–4763 (2004). Article CAS ADS Google
Scholar * Eftekharzadeh, B. et al. Sequence context influences the structure and aggregation behavior of a PolyQ tract. _Biophys. J._ 110, 2361–2366 (2016). Article CAS ADS Google
Scholar * Escobedo, A. et al. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. _Nat. Commun._ 10, 2034 (2019). Article ADS Google Scholar
* Lavery, D. N. & McEwan, I. J. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation
underlies structural plasticity and protein-induced folding. _Biochemistry_ 47, 3360–3369 (2008). Article CAS Google Scholar * De Mol, E. et al. EPI-001, a compound active against
castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. _ACS Chem. Biol._ 11, 2499–2505 (2016). Article Google Scholar * De Mol, E. et al. Regulation
of androgen receptor activity by transient interactions of its transactivation domain with general transcription regulators. _Structure_ 26, 145–152.e3 (2018). Article Google Scholar *
He, B., Kemppainen, J. A. & Wilson, E. M. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. _J. Biol. Chem._ 275,
22986–22994 (2000). Article CAS Google Scholar * He, B. et al. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor
activation function dominance. _Mol. Cell_ 16, 425–438 (2004). Article CAS Google Scholar * Qin, J., Vinogradova, O. & Gronenborn, A. M. Protein–protein interactions probed by nuclear
magnetic resonance spectroscopy. _Methods Enzymol._ 339, 377–389 (2001). * Dyson, H. J. & Wright, P. E. Unfolded proteins and protein folding studied by NMR. _Chem. Rev._ 104, 3607–3622
(2004). Article CAS Google Scholar * Tam, S. et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. _Nat. Struct. Mol.
Biol._ 16, 1279–1285 (2009). Article CAS Google Scholar * Monsellier, E., Redeker, V., Ruiz-Arlandis, G., Bousset, L. & Melki, R. Molecular interaction between the chaperone Hsc70 and
the N-terminal flank of huntingtin exon 1 modulates aggregation. _J. Biol. Chem._ 290, 2560–2576 (2015). Article CAS Google Scholar * Clerico, E. M., Tilitsky, J. M., Meng, W. &
Gierasch, L. M. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. _J. Mol. Biol._ 427, 1575–1588 (2015). Article CAS Google Scholar *
Schmid, D., Baici, A., Gehring, H. & Christen, P. Kinetics of molecular chaperone action. _Science_ 263, 971–973 (1994). Article CAS ADS Google Scholar * Karagöz, G. E. et al.
Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. _Cell_ 156, 963–974 (2014). Article Google Scholar * Rudiger, S., Schneider-Mergener, J. & Bukau, B. Its
substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. _EMBO J_. 20, 1042–1050 (2001). * Srinivasan, S. R., Gillies, A. T., Chang, L.,
Thompson, A. D. & Gestwicki, J. E. Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. _Mol. Biosyst._ 8, 2323–2333 (2012). Article
CAS Google Scholar * Rose, G. D., Geselowitz, A. R., Lesser, G. J., Lee, R. H. & Zehfus, M. H. Hydrophobicity of amino acid residues in globular proteins. _Science_ 229, 834–838
(1985). Article CAS ADS Google Scholar * Rüdiger, S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening
cellulose-bound peptide libraries. _EMBO J._ 16, 1501–1507 (1997). Article Google Scholar * Taylor, I. R. et al. The disorderly conduct of Hsc70 and its interaction with the Alzheimer’s
related Tau protein. _J. Biol. Chem._ 293, 10796–10809 (2018). Article CAS Google Scholar * Söderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by
proximity ligation. _Nat. Methods_ 3, 995–1000 (2006). Article Google Scholar * Zboray, L. et al. Preventing the androgen receptor n/c interaction delays disease onset in a mouse model of
SBMA. _Cell Rep._ 13, 2312–2323 (2015). Article CAS Google Scholar * Katsuno, M. et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and
bulbar muscular atrophy. _Neuron_ 35, 843–854 (2002). Article CAS Google Scholar * Takeyama, K.-I. et al. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen
receptor in Drosophila. _Neuron_ 35, 855–864 (2002). Article CAS Google Scholar * Chevalier-Larsen, E. S. et al. Castration restores function and neurofilament alterations of aged
symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. _J. Neurosci._ 24, 4778–4786 (2004). Article CAS Google Scholar * Yu, Z. et al. Androgen-dependent
pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. _J. Clin. Invest._ 116, 2663–2672 (2006). Article CAS Google Scholar * Fawzi, N.
L., Ying, J., Torchia, D. A. & Clore, G. M. Kinetics of amyloid beta monomer-to-oligomer exchange by NMR relaxation. _J. Am. Chem. Soc._ 132, 9948–9951 (2010). Article CAS Google
Scholar * Jayaraman, M. et al. Slow amyloid nucleation via α-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. _J. Mol. Biol._ 415, 881–899 (2012).
Article CAS Google Scholar * Wetzel, R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. _J. Mol. Biol_. 421, 466–490 (2012). * Young, Z. T. et al.
Stabilizing the Hsp70-Tau complex promotes turnover in models of tauopathy. _Cell Chem. Biol._ 23, 992–1001 (2016). Article CAS Google Scholar * Mayer, M. P. & Bukau, B. Hsp70
chaperones: cellular functions and molecular mechanism. _Cell. Mol. Life Sci._ 62, 670–684 (2005). Article CAS Google Scholar * Rousaki, A. et al. Allosteric drugs: the interaction of
antitumor compound MKT-077 with human Hsp70 chaperones. _J. Mol. Biol._ 411, 614–632 (2011). Article CAS Google Scholar * Walcott, J. L. & Merry, D. E. Ligand promotes intranuclear
inclusions in a novel cell model of spinal and bulbar muscular atrophy. _J. Biol. Chem._ 277, 50855–50859 (2002). Article CAS Google Scholar * Shao, H. et al. Exploration of benzothiazole
rhodacyanines as allosteric inhibitors of protein-protein interactions with heat shock protein 70 (Hsp70). _J. Med. Chem._ 61, 6163–6177 (2018). Article CAS Google Scholar * Yu, Z. et
al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. _Am. J. Pathol._ 168, 195–204
(2006). Article CAS Google Scholar * De Los Rios, P. & Barducci, A. Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. _Elife_ 3, e02218
(2014). Article Google Scholar * Cesa, L. C. et al. X-linked inhibitor of apoptosis protein (XIAP) is a client of heat shock protein 70 (Hsp70) and a biomarker of its inhibition. _J. Biol.
Chem._ 293, 2370–2380 (2018). Article CAS Google Scholar * Lieberman, A. P. Spinal and bulbar muscular atrophy. _Handb. Clin. Neurol._ 148, 625–632 (2018). Article Google Scholar *
Lieberman, A. P., Shakkottai, V. G. & Albin, R. L. Polyglutamine repeats in neurodegenerative diseases. _Annu. Rev. Pathol_. https://doi.org/10.1146/annurev-pathmechdis-012418-012857
(2018) * Pratt, W. B., Morishima, Y., Gestwicki, J. E., Lieberman, A. P. & Osawa, Y. A model in which heat shock protein 90 targets protein-folding clefts: rationale for a new approach
to neuroprotective treatment of protein folding diseases. _Exp. Biol. Med._ 239, 1405–1413 (2014). Article Google Scholar * Roth, D. M. et al. Modulation of the maladaptive stress response
to manage diseases of protein folding. _PLoS Biol._ 12, e1001998 (2014). Article Google Scholar * Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX
pipes. _J. Biomol. NMR_ 6, 277–293 (1995). Article CAS Google Scholar * Vranken, W. F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. _Proteins_ 59,
687–696 (2005). Article CAS Google Scholar * Weiss, A. et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models
of Huntington’s disease. _J. Neurochem._ 104, 846–858 (2008). CAS PubMed Google Scholar * Romero, P. et al. Sequence complexity of disordered protein. _Protein.: Struct. Funct. Bioinf._
42, 38–48 (2001). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS X.S. acknowledges funding from Obra Social “la Caixa”, AGAUR (2017 SGR 324), Marató TV3 (102030), MINECO
(BIO2012-31043 and BIO2015-70092-R) and the European Research Council (CONCERT, contract number 648201). IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from MINECO
(Government of Spain). A.P.L. acknowledges funding from NIH (NS055746). J.E.G. acknowledges funding from NIH (NS059690). S.R.N. was supported by a Rackham Predoctoral Fellowship and by the
NIH (GM007863). AUTHOR INFORMATION Author notes * These authors contributed equally: Bahareh Eftekharzadeh, Varuna C. Banduseela, Giulio Chiesa, Paula Martínez-Cristóbal. AUTHORS AND
AFFILIATIONS * Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028, Barcelona, Spain Bahareh Eftekharzadeh,
Giulio Chiesa, Paula Martínez-Cristóbal, Marta Marin-Argany, Claudio Di Sanza, Isabelle Brun-Heath, Jesús García, Ángel R. Nebreda & Xavier Salvatella * Joint BSC-IRB Research Programme
in Computational Biology, Baldiri Reixac 10, 08028, Barcelona, Spain Bahareh Eftekharzadeh, Giulio Chiesa, Paula Martínez-Cristóbal, Marta Marin-Argany, Claudio Di Sanza, Isabelle Brun-Heath
& Xavier Salvatella * Department of Pathology, University of Michigan, Ann Arbor, MI, 48109, USA Varuna C. Banduseela, Samir R. Nath, Elisa Giorgetti, Zhigang Yu & Andrew P.
Lieberman * University of California at San Francisco, Department of Pharmaceutical Chemistry, 675 Nelson Rising Lane, San Francisco, CA, 94158, USA Jennifer N. Rauch, Daniel M. C. Schwarz,
Hao Shao & Jason E. Gestwicki * CERM and Department of Chemistry “Ugo Schiff”, University of Florence, Via Luigi Sacconi 6, 50019, Sesto Fiorentino, Florence, Italy Roberta Pierattelli
& Isabella C. Felli * ICREA, Passeig Lluís Companys 23, 08010, Barcelona, Spain Ángel R. Nebreda & Xavier Salvatella Authors * Bahareh Eftekharzadeh View author publications You can
also search for this author inPubMed Google Scholar * Varuna C. Banduseela View author publications You can also search for this author inPubMed Google Scholar * Giulio Chiesa View author
publications You can also search for this author inPubMed Google Scholar * Paula Martínez-Cristóbal View author publications You can also search for this author inPubMed Google Scholar *
Jennifer N. Rauch View author publications You can also search for this author inPubMed Google Scholar * Samir R. Nath View author publications You can also search for this author inPubMed
Google Scholar * Daniel M. C. Schwarz View author publications You can also search for this author inPubMed Google Scholar * Hao Shao View author publications You can also search for this
author inPubMed Google Scholar * Marta Marin-Argany View author publications You can also search for this author inPubMed Google Scholar * Claudio Di Sanza View author publications You can
also search for this author inPubMed Google Scholar * Elisa Giorgetti View author publications You can also search for this author inPubMed Google Scholar * Zhigang Yu View author
publications You can also search for this author inPubMed Google Scholar * Roberta Pierattelli View author publications You can also search for this author inPubMed Google Scholar * Isabella
C. Felli View author publications You can also search for this author inPubMed Google Scholar * Isabelle Brun-Heath View author publications You can also search for this author inPubMed
Google Scholar * Jesús García View author publications You can also search for this author inPubMed Google Scholar * Ángel R. Nebreda View author publications You can also search for this
author inPubMed Google Scholar * Jason E. Gestwicki View author publications You can also search for this author inPubMed Google Scholar * Andrew P. Lieberman View author publications You
can also search for this author inPubMed Google Scholar * Xavier Salvatella View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS B.E., M.M.,
J.G, and X.S. carried out and analyzed NMR experiments. G.C. set up a protocol to express, purify, and study the aggregation of NTD1-155. J.N.R., D.M.C.S., and H.S. carried out FP and ITC
experiments. P.M-C. and C.D.S. carried out PLA, WB, and IF experiments. V.C.B., E.G., and S.R.N. analyzed effects of Hsp70-targeted small molecules in cell culture. V.C.B. and Z.Y. analyzed
effects of Hsp70-targeted molecules in mice. R.P., I.C.F., I.B-H. and A.N. contributed to the analysis and interpretation of the results. J.E.G., A.P.L. and X.S. conceived and led the
project and also wrote the first draft of the paper. All authors contributed to the final version. CORRESPONDING AUTHORS Correspondence to Jason E. Gestwicki, Andrew P. Lieberman or Xavier
Salvatella. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION: _Nature Communications_ thanks Lila Gierasch,
Harry T. Orr and the other anonymous reviewer for their contribution to the peer review of this work. Peer reviewer reports are available PUBLISHER’S NOTE: Springer Nature remains neutral
with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE REPORTING SUMMARY SOURCE DATA
SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution
and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if
changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the
material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to
obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Eftekharzadeh, B., Banduseela, V.C., Chiesa, G. _et al._ Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. _Nat
Commun_ 10, 3562 (2019). https://doi.org/10.1038/s41467-019-11594-y Download citation * Received: 04 January 2019 * Accepted: 23 July 2019 * Published: 08 August 2019 * DOI:
https://doi.org/10.1038/s41467-019-11594-y SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative