The structure of a red-shifted photosystem i reveals a red site in the core antenna
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Photosystem I coordinates more than 90 chlorophylls in its core antenna while achieving near perfect quantum efficiency. Low energy chlorophylls (also known as red chlorophylls)
residing in the antenna are important for energy transfer dynamics and yield, however, their precise location remained elusive. Here, we construct a chimeric Photosystem I complex in
_Synechocystis_ PCC 6803 that shows enhanced absorption in the red spectral region. We combine Cryo-EM and spectroscopy to determine the structure−function relationship in this red-shifted
Photosystem I complex. Determining the structure of this complex reveals the precise architecture of the low energy site as well as large scale structural heterogeneity which is probably
universal to all trimeric Photosystem I complexes. Identifying the structural elements that constitute red sites can expand the absorption spectrum of oxygenic photosynthetic and potentially
modulate light harvesting efficiency. SIMILAR CONTENT BEING VIEWED BY OTHERS STRUCTURE OF THE FAR-RED LIGHT UTILIZING PHOTOSYSTEM I OF _ACARYOCHLORIS MARINA_ Article Open access 20 April
2021 STRUCTURES OF A PHYCOBILISOME IN LIGHT-HARVESTING AND PHOTOPROTECTED STATES Article 31 August 2022 CRYO-EM STRUCTURE OF A FUNCTIONAL MONOMERIC PHOTOSYSTEM I FROM _THERMOSYNECHOCOCCUS
ELONGATUS_ REVEALS RED CHLOROPHYLL CLUSTER Article Open access 08 March 2021 INTRODUCTION Oxygenic photosynthesis, taking place in cyanobacteria, algae, and plants, powers the biosphere by
converting light photons into chemical energy. Photochemical conversion occurs in two photosystems, photosystem I and photosystem II (PSI and PSII respectively), which are multi-subunit
protein-pigment complexes located in thylakoid membranes. The cyanobacterial PSI is composed of a highly conserved core containing 11 subunits that coordinate 94–96 chlorophylls (Chls) and
22 carotenoids (Cars) per PSI monomer. These PSI monomers assemble into trimers or tetramers depending on the species1,2,3. Six of the PSI Chls, located at the center of the complex, make up
the internal electron transport chain. Two of these six Chls form a special pair called P700 which is oxidized to the relatively stable P700+ state (with a characteristic bleach around 700
nm) as part of the photochemically induced charge separation process in PSI. The rest of the Chls function as a core antenna for excitation energy transfer (EET). Altogether, PSI functions
with near perfect quantum efficiency; ~98% of the photons absorbed in the antenna result in photochemical charge separation in the reaction center4. One of the conserved features of PSI is
the existence of a small number of “red” or low-energy Chls, defined as Chls with an absorption maximum at longer wavelengths than P7005,6,7,8,9. The presence of red sites necessitates
uphill energy transfer steps as part of the function of PSI and this is why this group of Chls plays a major part in determining the rate of EET in the PSI antenna6,10,11,12. Specifically,
the trapping rate of the cyanobacterial PSI increases in rough proportion to its red sites content, from ~20 ps in complexes nearly devoid of red Chls to ~50 ps in PSI with an extremely
red-shifted Chls10,13. The large number of pigments in the core antenna and their similar electronic transitions makes the identification of these Chls extremely challenging12,14. In
solution, Chls can form dimers and higher order aggregates that result in a strong red spectral shift due to excitonic coupling15. It was suggested that Chl aggregates, dimers and trimers in
the PSI antenna, are the basic structural unit forming red sites in PSI antenna12,16,17. Several, not mutually exclusive functions were put forth with regards to the red Chls: It was
suggested that red Chls can focus the energy to P700 by reducing the number of EET steps between the bulk Chls and P70018. In addition, red Chls comprise up to 10% of the antenna Chls and
can contribute to light harvesting by absorbing light in the far-red region, a feature that is especially important in high-density cultures or shaded environments where light is not only
scarce but tend to be red shifted due to absorption by multiple cells19,20. Another suggestion is that red Chls play a role in photoprotection. It was shown that when P700 is oxidized and
the RC is closed, emission from the red sites is significantly reduced, potentially minimizing the formation of Chls triplets in PSI21,22. In plants, most of the red sites are located in the
peripheral antenna LHCI8. Cyanobacteria, which lack the LHCI antenna, carry their red sites in the core antenna of PSI. The properties and numbers of red Chls vary across different species:
estimations of the number of red Chls range from five in _Synechocystis_ PCC 6803 (_Synechocystis)_ to ten Chls in _Thermosynechococcus elongatus_ (_T. elongatus_)9. A major breakthrough in
identifying the location of red Chls was the elucidation of the PSI structure from T. _elongatus_1 which enabled the calculation of dipole interactions among the PSI Chls. This resulted in
the first assignment of red Chls in PSI internal antenna1. Since then, multiple attempts to model the steady state properties and energy transfer processes in PSI using various treatments
were presented23,24,25,26,27,28,29,30. However, very little structure−function analysis has been carried out on the core antenna and this is critical for every modeling attempt. Structures
of PSI with known differences in their red sites content are essential in order to reach the correct assignment of the red states in PSI2. From the Chls suspected as contributing to the red
states in _T. elongatus_, one unusual Chl trimer stands out as unique in PSI. This Chl trimer is made up of Chls B31-B32-B33 (Chl numbering follows1). The assignment of the B31-B32-B33
trimer as a red site is controversial and was disputed by some theoretical studies26,30. Different approaches attempted to fit several types of spectroscopic data with site energies values
assigned to the different PSI Chls. So far, the calculated site energies differ quite substantially between publications and in particular do not agree on the identity of red pigments in PSI
and their site energies23,25,27,28,30 (In fact, in most cases multiple sets of site energies can be used to fit the data). Overall, more than 30 different Chls were implicated as red in
various calculations with a consensus of only a single site (based on data from six different publications; Fig. 1a). Here we produced a chimeric PSI in cyanobacteria, introducing a small
loop which adds a single Chl (B33) to the PSI complex of _Synechocystis_, reconstituting the B31-B32-B33 Chl trimer from _T. elongatus_ in the _Synechocystis_ PSI. We demonstrate that this
additional Chl forms a new red state in _Synechocystis_ PSI (named C710), contributed to by the B31-B32-B33 trimer. We measure the effect of modifying a red state in PSI on the growth of
cells and the energy transfer processes in PSI. We solved the structure of the chimeric complex using single particle Cryo-EM and measured a new type of large-scale heterogeneity in the PSI
trimer, suggesting that each individual PSI monomer can occupy a range of positions in the PSI trimer. The location of low-energy sites in the core antenna and the large-scale heterogeneity
discovered in this study are universal properties of the cyanobacterial PSI. RESULTS INCORPORATION OF A NEW LOW-ENERGY CHLOROPHYLL INTO THE _SYNECHOCYSTIS_ PSI COMPLEX The structure of PSI
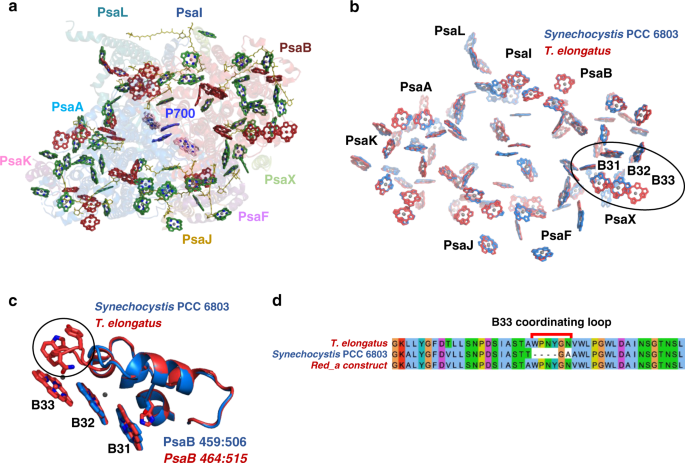
from _T. elongatus_ contains 96 Chl molecules coordinated by 12 protein subunits. All the different Chls suggested to contribute to the low-energy states in this system are colored in red in
Fig. 1a. Fig. 1b shows an overlay of the Chl networks from _T. elongatus_ and _Synechocystis_. _T. elongatus_ was shown to contain a larger number of red Chls at lower energies9. Most Chl
positions are absolutely conserved between _T. elongatus_ and _Synechocystis_. This similarity extends not only to the position and orientation of the Chl rings, but also to their
environments. One notable difference is an additional Chl in the _T. elongatus_ structure, B33. Examination of the protein environment in _T. elongatus_ PSI showed an additional loop
coordinating this Chl (Fig. 1c). Sequence alignment showed that this loop is located between conserved amino acids sequences. The extra loop sequence from _T. elongatus_ was inserted into
the _Synechocystis_ PsaB gene to form the Red_a PSI complex (Fig. 1d). ADDITION OF THE B33 CHL TO PSI INCREASES RED EMISSIONS IN CELLS Initial indications that Chl B33 contributes to a red
state in PSI were observed in cells. The fluorescence spectra of Red_a cells excited at the Chl Soret band (440 nm) showed a shoulder in the red region of the spectrum in comparison to the
wild type (WT; Fig. 2a). Examining the emission at 77 K showed that the main red emission peak, which originates from the PSI red states, is red shifted from 722 nm to 724 nm. Additional
peaks in the 77-K emission spectra, originating from PSII (684 nm and 694 nm) and phycobilisomes (648 nm), did not change their position, showing that only PSI was affected by the Red_a
mutation (Fig. 2b). We compared the growth of cells carrying the Red_a mutation at various light intensities to WT cells and strains carrying only the resistance gene and observed no
difference in the growth of cells over a wide range of light intensities showing that the Red_a PSI is functional in cells (Fig. 2c). The Red_a mutation did not affect the oligomeric state
of the purified PSI complex (Fig. 3a) as we observed the same trimer to monomer ratio in solubilized membranes. In addition, the same subunit composition is observed in both Red_a and WT
PSI, we conclude that the Red_a mutation does not interfere with the native structure of the complex (Fig. 3b). As seen in Fig. 3c, the absorption spectra of PSI complexes from WT and Red_a
strains overlap closely in the visible spectral region. The difference spectrum shows a major positive peak in the Qy region of PSI, centered ~709.3 nm. This clearly indicates that the added
B33 Chl contributes to one of the lowest-energy states in PSI and absorbs light at longer wavelengths than 700 nm (Fig. 3d). Gaussian approximation of the difference spectra was maximal at
~709.3 nm and shows a large full width at half maximum (fwhm) of ~25 nm (450 cm−1), this together with the intensity of the band suggests that more than one Chl molecule is involved, in
agreement with previous experiments suggesting that Chl aggregates constitute the red states in PSI. In the 350–520-nm region, a negative signal with a clear Car signature is observed. We
attribute this signal to a loosely bound carotene population present in the WT trimer. To test this, we subjected our samples to a further step of gentle purification, a repeat of the
sucrose gradient purification. This test showed that the difference in the carotene region is easily lost while the difference in the chlorophyll Qy region is nearly identical after this
extra purification step (Supplementary fig. 1c, d). The fluorescence spectra of the purified complexes at room temperature clearly show an increase in the Red_a complex emission in the red
part of the spectrum compared to WT (Fig. 3e), the differences between the WT and Red_a peaks near 720 nm. At 77 K, emission from the red states is clearly observed with a fluorescence band
maximum of 722 nm and 724 nm observed in the WT and Red_a, respectively (Fig. 3f). To get a more precise view of the differences between WT and Red_a, we measured the emission from both
complexes at 4 K (Supplementary fig. 2a). The results show that the emission contributed by the B31-B32-B33 Chl trimer (assigned below to the C710 state) peaks near 725 nm (at 4 K) and is
extremely broad (fwhm ~460 cm−1), suggesting a larger degree of mixing with the charge-transfer (CT) state(s) than that observed for the B31-B32 dimer present in WT PSI. LOW TEMPERATURE
STUDIES SHOW THAT B31-B32-B33 CONTRIBUTES TO THE C710 STATE IN PSI To minimize thermal broadening and get a clearer picture of the changes introduced into the Red_a mutant we carried out
low-temperature absorption and nonresonant hole burning (HB) measurements. Figure 4 shows the 4-K absorption spectra for the Red_a (red spectrum, curve A) and WT PSI (blue spectrum, curve
B). Both spectra were normalized in the Qy region to account for their Chl content. Similarly, to the observed differences in the room temperature absorption spectra, the absorption
difference between the Red_a mutant and WT PSI (within the lowest-energy band) accounts for an additional absorption of about two Chls per PSI monomer, not one Chl as expected in Red_a. The
difference between 4 K absorption spectra A and B revealed a positive band with a maximum near 710.4 nm with fwhm of ~13 nm (260 cm−1). Based on the Cryo-EM structure of the Red_a mutant
(see below), we can exclude that more than one Chl was incorporated per monomer. Therefore, we suggest that the oscillator strength of the low-energy states has increased due to intensity
stealing. Since the B31-B32 dimer in _Synechocystis_ was previously assigned to the C706 state31, we confirm that this state is indeed one of the low-energy states in WT PSI. Due to addition
of B33 Chl to the B31-B32 dimer, and increased interaction between the three Chls, the maximum of this state shifts to 710.4 nm. This new absorption band is assigned to the C710 state in
the reminder of the manuscript. That is, this state is contributed to by the B31-B32-B33 Chl trimer. Previously, HB measurements revealed the lowest-energy state in this system is actually
located at 714 nm (termed C714 trap)31. The nonresonant HB spectrum of the WT PSI complex (Fig. 4, curve c), shows the typical wide hole burned in this complex, centered at 714 nm, which
mostly represents the C714 state31. In contrast, in the Red_a PSI complex the nonresonant HB spectrum (Fig. 4, curve d) shows very broad hole with the minimum near 710 nm (indicated by the
asterisk in Fig. 4) and assigned to the C710 state. The broad hole includes contribution from the C714 state as well32, indicating that in the Red_a PSI complex, excitation energy is not
efficiently transferred to the lowest-energy C714 state at low temperatures. We anticipate that future frequency-dependent resonant hole-burning spectra obtained for several PSI mutants will
shed even more light on the EET dynamics in WT and mutated PSI complexes. Assignment of low-energy states (and their composition) in WT PSI and its Red_a mutant is summarized in Table 1.
TRAPPING IS AFFECTED IN THE RED_A MUTANT AT ROOM TEMPERATURE To monitor EET in PSI we measured the fluorescence decay kinetics of purified WT and Red_a mutant PSI trimers in the wavelength
region of 630–800 nm at room temperature using a streak camera setup. The overall emissions summed over the duration of the measured time window follows the general shape of the steady state
emission spectra, with Red_a showing enhanced emission in the red region of the emission profile (Fig. 5a). The fluorescence decay was fitted with three decay components using global
analysis33,34. Total decay summed over the 630–800-nm range is shown in Fig. 5b (complete data, fit, and residuals can be found in Supplementary fig. 3) and clearly show slower decay in the
Red_a PSI complex. Decay associated spectra (DAS) obtained from global analysis show a typical fast component with decay in the blue region of the spectrum (positive decay amplitudes) and a
rise component in the red region (negative decay amplitudes; Fig. 5c), originating from EET from the bulk antenna Chls to the red Chls as the core antenna equilibrates. The second decay
component is mostly associated with trapping processes and also includes contributions from bulk antenna Chls and the different red Chl pools (Fig. 5d). In addition, a low amplitude, slow
component associated with free pigments (Fig. 5e) is present in the data. While the fitted lifetimes of the fast components appear very different (4 and 8 ps in the WT and Red_a complex,
respectively) the differences should not be considered as significant as they are comparable to half of the instrument response function (IRF) of our setup (Fig. 5b, c). In addition,
Gaussian decomposition of the transfer components showed that the transfer processes from the major antenna to the red pools in WT and Red_a complexes are similar (Fig. 5c). In contrast, the
trapping component in Red_a is significantly slower than the WT trapping time (35 ps vs 28 ps). In addition, Gaussian decomposition of the DAS showed that the main differences between WT
and Red_a PSI complexes lay in the amplitudes of the 685–687-nm component which decreases significantly in Red_a. Accompanied with the blue band decrease, an increase of the 707–709 nm
component and a slight increase of the 717-nm component was observed, suggesting an excitation redistribution which is more towards the newly formed red states (B31-B32-B33) in Red_a. The
location of the B31-B32-B33 trimer within the Red_a PSI complex implies that trapping directly from the B31-B32-B33 trimer is unlikely. The Red_a “trapping” DAS must include an uphill energy
transfer to the bulk antenna at room temperature. Such process is extremely fast and is manifested via the decrease in the decay amplitude in the blue region of the trapping DAS together
with the rise in decay amplitudes in the red region of the trapping DAS. Altogether, large effect on trapping time is observed by the addition of a single Chl to the PSI antenna. C710
location, at the edge of the core antenna, where it is connected to a small number of adjacent Chls, probably contributes to this, as the necessary uphill transfer to the core antenna likely
proceeds through a limited number of pathways with currently unknown energetics. The sensitivity of the electronic structure of Chls to their environment implies that it is important to
measure the configuration of the newly formed B31-B32-B33 trimer in PSI in order to validate the structure of this trimer. In particular, B31-B32-B33 is in close proximity to PsaX, a PSI
subunit which is absent in _Synechocystis_ and this, together with additional sequence changes not included in the Red_a construct, can potentially affect the configuration of the Red_a
loop. CRYO-EM STRUCTURE OF THE RED_A PSI COMPLEX We determined the structure of the Red_a PSI trimer using Cryo-EM to a global resolution of 3.1 Å (Table 2 and Supplementary figs. 4 and 5).
The majority of the map contains information to 2.4 Å resolution (Supplementary fig. 5) and clearly shows pigment positions and orientations as well as side chains configurations with high
confidence (Fig. 6b, Supplementary fig. 6, and Supplementary table 1). Interestingly, we can account for all the Car molecules observed in the 2.5-Å crystal structure in spite of the
negative absorption difference in the Car region observed in Fig. 3d, consistent with this signal being associated with a loosely bound Car. As seen in Fig. 6c the added Chl, which is
located at the periphery of PSI, is clearly observed. We can also confidently model the loop coordinating this Chl and the conformation closely follows that of the _T. elongatus_ PSI
structure (Fig. 6c). We conclude that the added B33 Chl adopts the same configuration as in the _T. elongatus_ PSI structure with its dipole configuration parallel to B32, which is, in turn
parallel to B31. The closest distances between the Chls rings π systems over the trimer is ~3.7 Å, all of these factors contribute to the stronger electronic coupling between them leading to
the red-shifted C710 state. Altogether, we observe no additional structural changes between Red_a and the WT PSI, strongly supporting our claim that the addition of B33 is the underlying
cause for all of the observed spectroscopic differences. LARGE-SCALE STRUCTURAL HETEROGENEITY IN PSI The two PSI structures from _Synechocystis_, the WT trimer, obtained using X-ray
crystallography35 and the Red_a trimer, determined via Cryo-EM in this work, overlap closely over each of their PSI monomers (r.m.s.d of 0.56 Å). We can clearly resolve the 96 Chls (in
Red_a), 22 Car, and 9 lipid molecules present in both models. Interestingly, the overlaid trimeric structures revealed some variability between the relative positions of the PSI monomers
which is independent of the Red_a mutation. This is illustrated in Fig. 6d which shows both trimers superimposed on each other with one of the monomers (labeled ‘A’) used for alignment. It
is clearly seen that monomers ‘B’ and ‘C’ are shifted between the crystal and Cryo-EM structures. We measured the distance between each pairs of monomers and observed that distances between
the three PSI monomers vary by as much as 6 Å in the crystal structure. In contrast, inter monomer distances vary by only 0.1 Å in the Cryo-EM structure (Fig. 6d). These differences suggest
that the PSI trimer can exist in a range of conformations which should be observable as heterogeneity in our Cryo-EM dataset. We examined this aspect of the heterogeneity in our dataset
using multibody refinement, an implementation of focused refinement with iterative signal subtraction in Relion36,37. Multibody refinement consists of defining sub-volumes in the overall
map, in our case each PSI monomer was defined as a separate body using low-resolution masks (Supplementary fig. 7a). The orientation of each particle is then optimized to maximize the signal
in each sub-volume. Using this strategy, the refinement is free to converge on the different bodies even if their relative positions vary between the different single particle observations.
If successful, multibody refinement leads to improved maps and resolution estimates. The variability between the refined positional parameters can be used to measure the heterogeneity and
its direction in the particle dataset using principal component analysis36. Following multibody refinement of the Red_a dataset, the overall resolution decreased from 3.19 Å to 3. 1 Å
(Supplementary fig. 4). The estimated resolution at the peripheral regions of the map improved significantly (Supplementary fig. 7b). Most noticeably, the map around PsaK, a peripheral PSI
subunit, improved significantly showing better density around both protein and Chls (Supplementary fig. 7c). Altogether, the improved quality of the map and the appearance of chemically
correct features shows that our choice of segmenting the map to three PSI monomers is correct (other segmentation strategies were also tested). We next analyzed the heterogeneity in the
dataset using principal component analysis36. Supplementary figure 7d shows the eigenvalues of the 18 principal components for the data, where the first two components explain 36% of the
total variance in the data. By translating the refined maps along these principal components, it is possible to describe the heterogeneity in the data in the form of movies, where each frame
corresponds to 5% of the particle population along the specific component36. An example of the particle distribution along the first principle component is shown in Supplementary fig. 7e,
the unimodal distribution indicates that this heterogeneity is continuous and does not include distinct states. From Examining the movies generated from the first two principal components
(supplementary movies 1–6) showed that they describe very similar forms of heterogeneity depicted as side to side motions of the PSI monomers with the center of PSI, its trimerization
domain, serving as a hinge. The similar appearance of two principal components is possible because they involve different PSI monomers described by different parameters, nevertheless the
heterogeneity itself clearly belongs to a single class and accounts for more than a third of the variance across the dataset. To better model and measure this heterogeneity, we picked state
1 (representing the tail of the particle distribution along the first principal component containing 15% of the particles population) and state 2 (representing the opposite 15% of the
population) and refined the individual PSI monomers into these maps as rigid bodies. The resulting models are shown in Fig. 7a; we measured the distances between the two models at different
locations on the PSI trimer. These measurements clearly illustrate that monomers ‘B’ and ‘C’ move closer and further away in a circular fashion; the magnitude of the movement is proportional
to the distance from the center of the PSI trimer (Fig. 7a). Looking at the PSI trimer from the membrane plane, we measured similar shifts between state 1 and state 2 at the luminal and
stromal sides of the membrane, showing that the axis of the movement is nearly parallel to the membrane plane (Fig. 7a bottom and supplementary movies 3 and 4). The overall change in the
position of each PSI monomer is ~5 Å at the periphery of the trimer, however, since the monomers move towards and away from each other, the changes in the distances separating some of the
peripheral pigments are larger and can bring some of them to close proximity. This is shown in Fig. 7b, where closest Chl pairs between adjacent monomers in the different states were
measured. These Chls are coordinated by the PsaB subunit on one monomer and by the PsaA and PsaK subunits on the other. The average Mg to Mg distance between these clusters in the consensus,
averaged, Red_a structure is ~20 Å, significantly larger than the corresponding distances within the core antenna of each monomer which ranges between 9 Å and 13 Å1,2. In contrast, at the
state representing one conformation (between the ‘B’ and ‘C’ monomers) the average distances between these Chls drops to 18 Å, comparable with many antenna – PSI interfaces38,39,40. The Chls
configurations at the monomer’s interfaces are depicted in Fig. 7c with the distances from the closest state indicated. These distances are consistent with EET between monomers, especially
if we consider that they represent an upper bound to 15% of the PSI population. The orientation of one pair in this interface (B9–A20) is unfavorable for EET with an orientation factor value
of ~0.08 (Supplementary table 2 lists all the distances and orientation factors for the different states, similar calculation were described in refs. 38,41). However, other Chl pairs which
are closer together are favorably positioned for EET between monomers. We suggest that inter monomer EET through peripheral Chls is highly likely in a large fraction (approximately a third)
of the trimeric PSI population. To summarize, it is expected that large multimeric membrane complexes will exhibit some structural flexibility, especially when composed of large, complex
building blocks, like the PSI monomers. We are able to measure a form of structural heterogeneity in the PSI trimer of cyanobacteria which consists of large-scale shifts in the relative
monomer positions. While this heterogeneity is most easily displayed as movies, the time component in these movies cannot be established in Cryo-EM measurements and the time scale of the
transitions between the alternative conformations is unknown. Given the high conservation of PSI, we suggest this heterogeneity is a universal feature of PSI trimers. DISCUSSION The
existence of red Chls that absorb light in wavelengths longer than P700 in PSI has been known for close to 70 years42. The amount and spectral properties of the red Chls vary between
different PSI complexes and play a critical role in EET dynamics10. By introducing a structural feature into PSI we were able to incorporate a new Chl that contributed to a new red state and
extended the absorption spectrum of PSI. The spectroscopic changes derived from structural manipulation led to the first identification of a red site in cyanobacterial PSI internal antenna.
Spectral hole-burning and single-complex spectroscopies identified three red states in T. elongatus, C710, C715, and C71917,31. Assignment of the B31-B32-B33 Chl trimer to any one of these
low-energy states in _T. elongatus_ is controversial; for example, some studies assign C719, the lowest-energy state, to this trimer, based on modeling of time resolved data43, while other
studies assign one of the C710/C715 states to this Chl trimer44,45. Other studies did not assign a red state to the B31-B32-B33 trimer at all26,28,30. In contrast, our results clearly
support the assignment of a newly formed C710 state to the B31-B32-B33 trimer in the _Synechocystis_ Red_a PSI complex, and by inference suggest that C710 in _T. elongatus_ should be
assigned to the B31-B32-B33 trimer. This is based on both the differences in absorbance observed in Red_a and the formation of a new hole in the non-photochemical HB spectra at 710 nm
observed in Red_a (Figs. 3d and 4). It should be noted that while the conformations of the Chl rings in the newly formed B31-B32-B33 trimer in Red_a are highly similar to the Chl trimer in
_T. elongatus_, the Chl coordination of B31 is not identical in both species and was not replaced in Red_a, leaving room for additional differences. Previously, the B31-B32 dimer in the WT
_Synechocystis_ was assigned the C706 state31. This, together with our assignment of the C710 state to B31-B32-B33, suggest that the low-energy property of this site within the core PSI
antenna is conserved and the variation that is observed in red sites reflects modulation of site energy rather than changes in the location of the low-energy sites in PSI. Recently
additional PSI structures became available. In all currently known cyanobacterial PSI structures, there is a correlation between the presence of the Red_a loop, the coordination of a Chl
trimer and additional red absorbance. The red loop we added to the _Synechocystis_ WT PSI based on _T. elongatus_ structure can also be found in _Thermosynechococcus vulcanus_46,
_Fischerella thermalis_47 and _Nostoc_ sp. PCC 712048,49. The short loop present in _Synechocystis_ can be found in _Synechococcus_ sp. PCC 794250 and _Chroococcidiopsis_ sp. TS-821 (PDBID:
6QWJ)51. The absence of the red loop in those structures leaves a Chl dimer in this location (which should correspond to the C706 state). The Red_a loop can be found in different forms of
PSI, from thermophilic and mesophilic organisms and trimeric and tetrameric oligomeric states of PSI. This loop is a modular chlorophyll coordinator and we suggest that adding this loop to
any cyanobacterial PSI lacking this loop will lead to the formation of a B31-B32-B33-like Chl trimer resulting in addition, red shifted, absorption. The PSI complex from _Synechococcus_ WH
7803 was suggested to contains no red Chls13, Inspecting the PsaB sequence from _Synechococcus_ WH 7803 revealed that the B32 coordinating region is different than either _Synechocystis_ or
_T. elongatus_. Suggesting that the B32 Chl is either displaced or absent, excluding any coupling that exist between B31-B32 in _Synechocystis_ and resulting in the absence of the C706 state
in the _Synechococcus_ WH 7803 PSI complex. We propose a red site assignment in Table 1 which incorporates the findings from the current work and from several other publications (see table
for details). The presence of red Chls in the antenna of PSI necessitates an uphill energy transfer with thermal energy bridging the energetic gap between the red sites and P700 under
physiological temperatures. In Gobets et al.10, a unified minimalistic compartmental model was used to fit kinetic measurements from several cyanobacterial species, concluding that two
different red Chl pools, differing in their connectivity to P700, exist in PSI from _T. elongatus_ (and _Arthrospira platensis_) compared to a single red pool in _Synechocystis_. The
location of the B31-B32-B33 Chl trimer compared to other proposed red sites (see Table 1 and Fig. 8) which are significantly closer to P700, suggest that it makes one pool, separate from
other red sites. We suggest that a partial explanation for the relatively large effect of the Red_a mutation on trapping time is attributed to its location. Preliminary calculations suggest
that ~4 nm red spectral shift are obtained upon the addition of Chl B33 to the B31-B32 dimer, suggesting that this dimer should be assigned to the C706 state, in agreement with literature
data52. Thus, at least two red Chls pools exist in this cyanobacterium in both WT and Red_a. Recently, 2DES was applied to PSI from _Synechocystis_ and also detected two processes associated
with its red Chls, concluding that two red pools exist in this system52. It was suggested that red Chls can focus excitation energy to P700, contribute to light harvesting and participate
in photoprotection. Clearly, for the B31-B32-B33 Chl trimer focusing excitation energy to P700 is not a plausible suggestion. However, B31-B32-B33 may play a role in EET to PSI from the IsiA
antenna. The recent structure of PSI-IsiA shows that the predicted terminal emitters of IsiA are located in close proximity to the Red_a site suggesting a role in EET from IsiA to PSI38.
The B31-B32-B33 trimer also contributes to light harvesting in the far-red region of the spectrum, in spite of the increase in trapping time, the quantum efficiency of PSI remains very high.
It is also possible that B31-B32-B33 Chl trimer participates in photoprotection. The location of Red_a makes a direct interaction with P700 unlikely, however it is interesting to envisage a
role of red sites before the assembly of the PsaA-PsaB heterodimer and the formation of P700 itself. In the whole complex, P700 functions as a quencher of the PSI Chls, yet in the assembly
process of PSI, PsaB (in _C. reinhardtii_, for example) is synthesized first and integrated into the thylakoid membrane before PsaA53. The assembly process of PSI in cyanobacteria is still
unclear, however there is evidence for separate transcripts for PsaA and PsaB in addition to a whole transcript of PsaAB in _Synechocystis_ implying the possibility of similar assembly
mechanism as in _C. reinhardtii_54,55. P700 is created only upon dimerization of PsaA and PsaB and the formation of the whole internal electron transport chain of PSI is completed with the
subsequent addition of the stromal subunit, PsaC. PsaA and PsaB already carry Chls in their internal antenna that are prone to damage without the presence of the energy quencher, P700. In
this case, red Chls can be a part of a mechanism for energy quenching to prevent Chls triplet formation and damage to each subunit. The reason that we find this red site on PsaB is due to
the role of PsaB as an anchor for PsaA during PSI assembly. Each PSI monomer coordinates 22 Car molecules across the antenna which are known to play a crucial role in photoprotection. These
Cars interact with approximately two thirds of the antenna Chls, close examination shows that a Car is located in the vicinity of each of the predicted red sites in PSI (Fig. 8). The close
interaction between red Chls and Cars may be an important mechanism for photoprotection in PSI. The large-scale heterogeneity we observe occurs in the trimeric form of PSI and is independent
of the Red_a mutation. Previously, the red states in the cyanobacterial PSI were also shown to change significantly between trimeric and monomeric PSI forms, with red absorption
significantly decreased in monomers compared to trimers32,56. The reasons for these changes can be either conformational changes that modulate site energy or loss of pigments that occur upon
monomerization. We modeled the heterogeneity in PSI in the form of rigid monomers which cannot detect any conformational changes within the PSI monomers. However, it seems very reasonable
that conformational changes occur as part of the different states the monomers occupy, the extent of these conformational changes could affect the energy of a red site. One clear candidate
for this is the site formed by the B7-A31-A32 cluster assigned to the C707 state (see Table 1 and Fig. 8), which is in close proximity to the PsaL subunit known to be essential for
trimerization. Another clear consequence of the observed states are the large changes in distances between two groups of Chls located at the “corners” of PSI (Fig. 7). These changes enable
EET between the different PSI monomers. One of the proposed functions of the trimeric organization of PSI in cyanobacteria is to enable EET between monomers in the trimers. This was
suggested to relieve excess energy burden from one monomer by the adjacent monomer and slower rates of P700 oxidation were observed in the trimer of _Arthrospira platensis_ in comparison to
the monomer, suggesting energy migration between monomers57. The very same group of peripheral Chls highlighted in Fig. 7 was proposed to mediate this EET, however, large scale changes in
monomer packaging had to be assumed58. The heterogeneity identified in the current study means that a substantial portion of the trimeric PSI population occupies positions compatible with
monomer to monomer EET, although the nature of the heterogeneity is very different than the one proposed in58. PSI is one of the essential components of oxygenic photosynthesis. The high
density of pigments in the core of PSI presents a great challenge to deciphering the structural determinants of its electronic structure. The identification of the B31-B32-B33 trimer as one
of the red states brings us a step closer to understanding the design rules that drive the evolution of photosystems and allow these assemblies to tune their light-harvesting properties.
METHODS STRAIN CONSTRUCTION AND GROWTH _Red_a strain construction_: a plasmid (p60) containing the entire PsaAB operon marked with Kanamycin resistance gene 0.8Kb downstream to PsaB was
constructed from four PCR amplified fragments. The PsaAB operon and PsaB downstream fragments were amplified using the PsaAB_fwd/PsaAB_rev and PsaBdown_fwd/PsaBdown_rev primer pairs from the
_Synechocystis_ sp. PCC6803 genome. The pA15 Origin of replication was amplified from pACYC184 using primers pA15ori_fwd and pA15ori_rev. The kanamycin resistance gene was amplified from
pET30 using Kan_fwd and Kan_rev. The four fragments were assembled using the NEBuilder® HiFi DNA Assembly Master Mix. The Red_a mutation was constructed into p60 by adding the red loop
sequence using the Red_a fragment1 and fragment2 primers and p60 as a template. The two fragments were assembled using the NEBuilder® HiFi DNA Assembly Master Mix. All plasmids were
sequenced before being used to transform _Synechocystis_ sp. PCC6803 according to standard protocols. Complete and correct replacement of PsaB was verified by PCR and sequencing. The primers
used to construct the Red_a mutant are listed in supplementary table 3. _Culture conditions_: cyanobacteria were cultured in BG11 medium supplemented with 6 µg/ml Ferric ammonium citrate, 5
mM glucose, and 10 µg/ml Kanamycin under continuous white light (∼40 µE) in 30 °C. _Growth tests_: wild type, mutant, and control cells (containing the resistance gene but not the mutation)
were cultured in BG11 liquid medium supplemented with 6 µg/ml Ferric ammonium citrate and 5 mM glucose under continuous white light (∼40 µE) in 30 °C. The optical density was adjusted to
0.5 at 730 nm. Each culture was diluted x5, x25, x125, and x625 and grew on BG11 plates in different light intensities. _Photosystem I purification_: 20 l of cells were grown in BG11
(supplemented with 6 µg/ml ferric ammonium citrate and 5 mM glucose, plus 10 µg/ml kanamycin for the red strain) under light intensity of ∼40 µE at 30 °C. Cells were harvested using
centrifugation in a F9 6 × 1000 LEX rotor in Sorval LYNX 6000 centrifuge (Thermo Scientific) for 5 min, 4700 × _g_, all further sample handling was done on ice. Cells were washed once using
~200 ml of STN1 buffer (30 mM Tricine-NaOH pH 8, 15 mM NaCl, and 0.4 M sucrose). Spun down again using a F20-12 × 50 LEX rotor, 5 min, 12,857 × _g_. Finally, cells were resuspended in 50 ml
of STN1, and broken by Constant Systems cell disruptor (two cycles at 30 kpsi). The lysate was cleared by centrifugation in a F20-12 × 50 LEX rotor for 10 min at 18,514 × _g_. Membranes in
the supernatant were pelleted using ultracentrifugation (Ti70 rotor, 207,870 × _g_ for 2 h), and resuspended in 50 ml STN2 (30 mM Tricine-NaOH pH 8, 150 mM NaCl, 0.4 M sucrose). After
resuspension in STN2, the membranes were incubated on ice for 30 min, then collected again (Ti70 rotor, 207,870 × _g_, 2 h), and resuspended in ~15 ml of STN1. Chlorophyll concentration was
determined according to Amon59. n-Dodecyl β-d-maltoside (DDM, Glycon) was added to the membranes at a 15:1 DDM to chlorophyll mass ratio. The suspension was gently mixed by hand a few times
then incubated on ice for 30 min. After solubilization, the insoluble material was discarded using ultracentrifugation (Ti70, 207,870 × _g_, 30 min). The solubilized membranes were loaded
onto a DEAE gravity flow column (Toyopearl C650). The complexes were eluted using a linear NaCl gradient (15–350 mM NaCl) in 30 mM Tricine-NaOH pH 8, 0.2% DDM. Dark green fractions were
collected, PEG3350 (Hampton research) was added to a final concentration of 8.5% and the PSI trimer were precipitated using a 5 min, 3214 × _g_ spin in a F20-12 × 50 LEX rotor. The green
precipitate was resuspended in 30 mM Tricine-NaOH, pH 8, 0.05% DDM, 75 mM NaCl, and loaded onto a 10–30% sucrose density gradient, prepared in 30 mM Tricine-NaOH pH 8, 0.05% DDM, 75 mM NaCl.
Following centrifugation (SW40 rotor, 242,832 × _g_, 12 h), the PSI trimer was collected and precipitated with 8.5% PEG3350 at 150 mM NaCl. After a centrifugation step (18,407 × _g_, 5 min)
the green precipitate was resuspended in 30 mM Tricine-NaOH, pH 8, 0.05% DDM, 75 mM NaCl, and loaded onto a second 10–30% sucrose density gradient done in an SW60 rotor (392,960 × _g_, 4
h). The green band was collected and used for subsequent experiments. For the experiments using a third sucrose gradient (Supplementary fig. 1), the sample was diluted to 50% using 30 mM
Tricine-NaOH, pH 8, 0.05% DDM, 75 mM NaCl, and loaded on a 10–30% sucrose density gradient prepared as before (SW40 rotor, 242,832 × _g_, 12 h). ABSORPTION AND FLUORESCENCE SPECTROSCOPY
_Room temperature measurements_: absorption spectra were recorded on a Cary 4000 UV–Vis spectrophotometer (Agilent Technologies). Fluorescence spectra were recorded on a Fluoromax-4
spectrofluorometer (HORIBA Jobin-Yvon). Samples were diluted to an optical density of 0.8 and 0.1 at 680 nm for absorption and fluorescence measurements respectively, using buffer containing
30 mM Tricine-NaOH pH 8, 15 mM NaCl, and 0.05% DDM. The resulting spectra were normalized to the area of the chlorophyll Q bands (550–750 nm) using 95 chlorophylls for the wild type and 96
chlorophylls for Red_a. For 77 K fluorescence measurement samples were adjusted to an OD680 of 0.1 in a buffer of 50% glycerol 30 mM Tricine pH = 8.0, 15 mM NaCl, and 0.02% DDM. An Oxford
instruments Cryostat was used to cool the sample to 77 K (cells were plunged into liquid nitrogen and measured immersed in liquid nitrogen). Figures were prepared using OriginPro
(OriginLab). _Low-temperature absorbance_: for low-temperature experiments, samples were diluted with 1:2 (v/v) buffer:glass solution. The glass forming solution was 55:45 (v/v)
glycerol:ethylene glycol. Briefly, a Bruker HR125 Fourier transform spectrometer was used to measure the absorption and nonresonant hole-burned (NRHB) spectra with resolution of 4 cm−1. In
all, 488.0 nm excitation for NRHB spectra was produced from a Coherent Innova 200 argon ion laser. In total, 670.0 nm excitation was obtained from a Coherent CR899 ring dye laser pumped by a
Spectra-Physics Millennia Xs diode laser (532 nm). With laser dye LD 688 (Exciton), laser excitation in the spectral range of 650–720 nm (for resonantly burned holes) was available with a
line width 0.07 cm–1. Laser power in all experiments was precisely set by a continuously adjustable neutral density filter. Low temperature (4 K) experiments were performed using an Oxford
Instruments Optistat CF2 cryostat with sample temperature read and controlled by a Mercury iTC temperature controller. The hole-burning (HB) spectroscopy relies on differences observed in
the absorption spectrum of a low-temperature system after narrow-band laser excitation. If a pigment molecule (in resonance with the laser) experiences photochemical reaction, it ceases to
absorb at its original wavelength/frequency and one speaks of photochemical HB (PHB). In this work, however, we use non-photochemical HB (NPHB); in this case, the pigment molecule does not
undergo a chemical reaction, but its immediate environment experiences rearrangement on the protein energy landscape3,4. NPHB may result in the formation of persistent holes, meaning the
holes persist after the initial excitation is turned off, as long as low temperature is maintained. (The HB spectrum is obtained as the difference between the measured absorption spectrum
before and after laser excitation). The key information provided by HB spectroscopy (relevant to data discussed in this work) includes lifetimes of the zero-point level of S1(Qy)-states due
to EET, as determined by the widths of zero-phonon holes (ZPHs) and/or electron-phonon (protein) coupling parameters. _Time resolved fluorescence decay_: samples were diluted to an optical
density (680 nm) of 0.75 in 30 mM Tricine pH 8.0, 15 mM NaCl, 0.05% DDM, 5 mM Ascorbic acid, and 1.5 µM phenazine methosulfate. The excitation beam was set to excite the sample as close as
possible to the outer edge of the cuvette to reduce reabsorption effects. Experiments were carried out at room temperature in a 10.00 × 10.00 mm quartz cuvette (Hellma Analytics)
continuously mixed using a magnetic stirrer. Sample were illuminated by 100 fs pulses from a mode-locked Ti:S laser (Mira 900, Coherent Laser Inc., Santa Clara, CA) pumped by a
frequency-doubled Nd:YVO 4 laser (44% from an 18 W Verdi, Coherent Laser Inc.). The repetition rate was reduced to 4.75 MHz using a pulse picker (Model 9200, Coherent Laser Inc.) and the
excitation light was frequency-doubled to 400 nm and focused on the sample cuvette. To avoid singlet-singlet annihilation, the pulse energy was reduced to ~0.1 nJ using a neutral density
filter. Fluorescence was collected at a right angle to the excitation beam and focused on the entrance slit of a Chromex 250IS spectrograph coupled to a Hamamatsu C5680 streak camera with a
M56757 sweep unit. Time-intensity surfaces were recorded at a time scale of 0.8 ns on a Hamamatsu C4742 CCD camera. The fwhm of the overall instrument response function (IRF) was ~12 ps at
the 0.8 ns timescale. Global analysis was carried out in MATLAB using a set of scripts available from the author upon request following the methodologies described in33,34. CRYOGENIC
ELECTRON MICROSCOPY _Sample preparation for single particle Cryo-EM analysis_: the PSI band from the sucrose gradient was collected, NaCl was added to a final concentration of 150 mM and the
complex was precipitated using 8.5% PEG3350. After centrifugation (2,300 × _g_, 5 min in an Eppendorf tabletop), the green precipitate was resuspended in buffer (30 mM Tricine-NaOH pH 8,
150 mM NaCl 0.02% DDM), and any undissolved material was removed by repeating the centrifugation step (18,407 × _g_, 5 min). The chlorophyll concentration in the soluble material was
adjusted to 1 mg/ml using the above buffer. A 3 µl of The PSI complex was added to a holey carbon grids (C-flat 1.2/1.3 Cu 400 mesh grids (Protochips, Raleigh, NC)) after soaking the grids
in buffer. The sample was vitrified by flash plunging the grid into liquid ethane using manual plunger with blotting time of ~6 s. The grids were stored in liquid nitrogen until data
collection. _Data acquisition_: the Cryo-EM specimens were imaged on a Titan Krios transmission electron microscope (Thermo Fisher - FEI, Hillsboro, OR). The electron images were recorded
using a K2 Summit direct electron detect (DED) camera (Gatan, Pleasanton, CA) at super-resolution counting mode. Automated data collection was with SerialEM60 with stage shift in between
exposures. The defocus was set to vary between −1.5 and −3 μm. The super-resolution pixel size was 0.525 Å at the specimen level corresponding to ×47,600 magnification. The counting rate was
adjusted to 5.6 e−/Å2sec. Total exposure time was 8 s, fractionated into 40 frames, accumulating to a dose of 44.8 e−/Å2. _Data processing_: a flow chart describing data handling is shown
in Supplementary fig. 4. MotionCor261 was used to register the translation of each sub-frame, and the generated averages were binned 2X and dose-weighted62. Contrast Transfer Function (CTF)
parameters for each corrected micrograph were determined using CTFFIND463. Relion3.1 was used for the subsequent data processing37. A set of manually picked particles (~400) was subjected to
a few rounds of unsupervised 2D classification. Four class averages, which represented different orientations of the expected particle, were selected and used as the templates for the
automated particle picking procedure as implemented in Relion which yielded 927,010 particles. This particle set was subjected to several rounds of unsupervised 2D classification leading to
a set of 231,381 particles which were extracted as boxes of 310 pixels with a final pixel size of 1.05 Å/Px. 3D reconstruction using this set led to a map with an overall resolution of 3.9 Å
after masking. This particle set was subjected to 3D classification using six classes with most of the particles classified in clear PSI trimer classes and minor fraction classified into a
“junk” class, the trimeric classes were grouped together and refined again and then the 3D classification was rerun (three cycles of classification followed by refinement) until only trimer
classes were obtained. All classes were judged to be similar by visual inspection of the maps and all refined to comparable resolution (~4 Å). At this point all the classes were combined to
yield a 191,798 particle set. Per particle CTF refinement was carried out using relion3.164 followed by Bayesian polishing65. The final resolution of this particle set refined in C1 was 3.35
Å. After this refinement the particle set was symmetry expanded using the C3 group and refined to a final resolution of 3.19 Å. Finally, multibody refinement36 using each monomer as a
separate body yielded the final map at a resolution of 3.1 Å according to gold standard FSC criteria66. The final map was sharpened using the post processing procedure in Relion, the
b-factor used for sharpening was −100 Å2. Local resolution was estimated using ResMap67. The three PSI monomers were refined into each of the three maps as rigid bodies and a composite map
was generated in phenix68 using the combine_focused_maps tool. _Model building and refinement_: the initial PSI model was taken from the 2.5-Å x-ray structure of the trimeric PSI from
_Synechocystis_ (PDBID: 5OY0)35 with PsaK taken from69 (PDBID: 6KIG) and the sequence was manually changed to the _Synechocystis_ sequence. The model was docked into the map using PHENIX.
The final model was refined against the Cryo-EM density map using phenix.real_space_refine70,71. Final model statistics are shown in Table 1. Map model fitting and resolution was also
checked using MapQ, a USCF Chimera plugin72. PyMOL73 and UCSF Chimera74 were used to generate all images. Dipole orientations were calculated using an R script similarly to38,41. REPORTING
SUMMARY Further information on research design is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The final model (PDB 6UZV) and map (EMD 20963)
were deposited in the Protein Databank and Electron Microscopy Database, respectively. All other data are available from the authors upon reasonable request. Source data are provided with
this paper. CODE AVAILABILITY Global analysis of fluorescence data was carried out using a MATLAB script, dipole orientations values were calculated using an R script, both available from
the author upon reasonable request. CHANGE HISTORY * _ 20 NOVEMBER 2020 A Correction to this paper has been published: https://doi.org/10.1038/s41467-020-19953-w _ REFERENCES * Jordan, P. et
al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. _Nature_ 411, 909–917 (2001). Article ADS CAS PubMed Google Scholar * Mazor, Y., Nataf, D.,
Toporik, H. & Nelson, N. Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803. _Elife_ 3, e01496 (2013). Article PubMed
Google Scholar * Li, M. et al. Physiological and evolutionary implications of tetrameric photosystem I in cyanobacteria. _Nat. Plants_. https://doi.org/10.1038/s41477-019-0566-x (2019). *
Nelson, N. & Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. _Annu. Rev. Biochem._ 84, 659–683 (2015). Article CAS PubMed Google Scholar * Giera,
W. et al. Uphill energy transfer in photosystem I from Chlamydomonas reinhardtii. Time-resolved fluorescence measurements at 77 K. _Photosynth. Res._ 137, 321–335 (2018). Article CAS
PubMed Google Scholar * Shubin, V. V., Bezsmertnaya, I. N. & Karapetyan, N. V. Isolation from Spirulina membranes of two photosystem I-type complexes, one of which contains chlorophyll
responsible for the 77 K fluorescence band at 760 nm. _FEBS Lett._ 309, 340–342 (1992). Article CAS PubMed Google Scholar * Palsson, L. O., Dekker, J. P., Schlodder, E., Monshouwer, R.
& Van Grondelle, R. Polarized site-selective fluorescence spectroscopy of the long- wavelength emitting chlorophylls in isolated Photosystem I particles of Synechococcus elongatus.
_Photosynth. Res_. https://doi.org/10.1007/BF00041014 (1996). * Croce, R., Zucchelli, G., Garlaschi, F. M. & Jennings, R. C. A thermal broadening study of the antenna chlorophylls in
PSI- 200, LHCI, and PSI core. _Biochemistry_. https://doi.org/10.1021/bi9813227 (1998). * Karapetyan, N. V. et al. Long-wavelength chlorophylls in photosystem I of cyanobacteria: origin,
localization, and functions. _Biochemistry_ 79, 213–220 (2014). CAS PubMed Google Scholar * Gobets, B. et al. Time-resolved fluorescence emission measurements of photosystem I particles
of various cyanobacteria: a unified compartmental model. _Biophys. J._ 81, 407–424 (2001). Article ADS CAS PubMed PubMed Central Google Scholar * Pålsson, L.-O. et al. Energy transfer
and charge separation in photosystem I: P700 oxidation upon selective excitation of the long-wavelength antenna chlorophylls of Synechococcus elongatus. _Biophys. J._ 74, 2611–2622 (1998).
Article ADS PubMed PubMed Central Google Scholar * Gobets, B. et al. Polarized site-selected fluorescence spectroscopy of isolated Photosystem I particles. _Biochim. Biophys. Acta
Bioenerg._ 1188, 75–85 (1994). Article CAS Google Scholar * Van Stokkum, I. H. M. M. et al. Energy transfer and trapping in red-chlorophyll-free photosystem i from Synechococcus WH 7803.
_J. Phys. Chem. B_ 117, 11176–11183 (2013). Article PubMed CAS Google Scholar * van der Lee, J. et al. Steady-state polarized light spectroscopy of isolated Photosystem I complexes.
_Photosynth. Res_. https://doi.org/10.1007/BF00016562 (1993). * Shipman, L. L., Cotton, T. M., Norris, J. R. & Katz, J. J. An analysis of the visible absorption spectrum of chlorophyll a
monomer, dimer,and oligomers in solution. _J. Am. Chem. Soc._ 98, 8222–8230 (1976). Article CAS PubMed Google Scholar * Hayes, J. M., Matsuzaki, S., Rätsep, M. & Small, G. J. Red
chlorophyll a antenna states of photosystem I of the Cyanobacterium Synechocystis sp. PCC 6803. _J. Phys. Chem. B_ 104, 5625–5633 (2000). Article CAS Google Scholar * Zazubovich, V. et
al. Red antenna states of photosystem I from cyanobacterium Synechococcus elongatus: a spectral hole burning study. _Chem. Phys._ 275, 47–59 (2002). Article CAS Google Scholar * Valkunas,
L., Liuolia, V., Dekker, J. P. & van Grondelle, R. Description of energy migration and trapping in photosystem I by a model with two distance scaling parameters. _Photosynth. Res_.
https://doi.org/10.1007/BF00042972 (1995). * Trissl, H.-W. Long-wavelength absorbing antenna pigments and heterogeneous absorption bands concentrate excitons and increase absorption cross
section. _Photosynth. Res_. 35, 247–263 (1993). Article CAS PubMed Google Scholar * Rivadossi, A., Zucchelli, G., Garlaschi, F. M. & Jennings, R. C. The importance of PS I
chlorophyll red forms in light-harvesting by leaves. _Photosynth. Res._ 60, 209–215 (1999). Article CAS Google Scholar * Schlodder, E., Çetin, M., Byrdin, M., Terekhova, I. V. &
Karapetyan, N. V. P700 +- and 3P700-induced quenching of the fluorescence at 760 nm in trimeric Photosystem I complexes from the cyanobacterium Arthrospira platensis. _Biochim. Biophys. Acta
Bioenerg_. https://doi.org/10.1016/j.bbabio.2004.08.009 (2005). * Herascu, N. et al. Spectral hole burning in cyanobacterial photosystem i with P700 in oxidized and neutral states. _J.
Phys. Chem. B_ 120, 10483–10495 (2016). Article CAS PubMed Google Scholar * Yang, M. et al. Energy transfer in photosystem I of cyanobacteria Synechococcus elongatus: Model study with
structure-based semi-empirical Hamiltonian and experimental spectral density. _Biophys. J._ 85, 140–158 (2003). Article ADS CAS PubMed PubMed Central Google Scholar * Brüggemann, B.,
Sznee, K., Novoderezhkin, V., van Grondelle, R. & May, V. From structure to dynamics: modeling exciton dynamics in the photosynthetic antenna PS1. _J. Phys. Chem. B_ 108, 13536–13546
(2004). Article CAS Google Scholar * Şener, M. K. et al. Excitation migration in trimeric cyanobacterial photosystem I. _J. Chem. Phys._ 120, 11183–11195 (2004). Article ADS PubMed CAS
Google Scholar * Sener, M. K. et al. Robustness and optimality of light harvesting in cyanobacterial photosystem I. _J. Phys. Chem. B_. https://doi.org/10.1021/jp020708v (2002). * Byrdin,
M. et al. Light harvesting in photosystem I: modeling based on the 2.5-A structure of photosystem I from Synechococcus elongatus. _Biophys. J._ 83, 433–457 (2002). Article ADS CAS PubMed
PubMed Central Google Scholar * Damjanović, A., Vaswani, H. M., Fromme, P. & Fleming, G. R. Chlorophyll excitations in photosystem I of Synechococcus elongatus. _J. Phys. Chem. B_
106, 10251–10262 (2002). Article CAS Google Scholar * Kramer, T. et al. Energy flow in the Photosystem I supercomplex: Comparison of approximative theories with DM-HEOM. _Chem. Phys_.
https://doi.org/10.1016/j.chemphys.2018.05.028 (2018). * Adolphs, J., Muh, F., Madjet, M. E.-A., am Busch, M. S. & Renger, T. Structure-based calculations of optical spectra of
photosystem I suggest an asymmetric light-harvesting process. _J. Am. Chem. Soc._ 132, 3331–3343 (2010). Article CAS PubMed Google Scholar * Riley, K. J., Reinot, T., Jankowiak, R.,
Fromme, P. & Zazubovich, V. Red antenna states of photosystem i from cyanobacteria Synechocystis PCC 6803 and Thermosynechococcus elongatus: Single-complex spectroscopy and spectral
hole-burning study. _J. Phys. Chem. B_ 111, 286–292 (2007). Article CAS PubMed Google Scholar * Rätsep, M., Johnson, T. W. W., Chitnis, P. R. R. & Small, G. J. J. The red-absorbing
chlorophyll a antenna states of photosystem I: a hole-burning study of Synechocystis sp. PCC 6803 and its mutants. _J. Phys. Chem. B_ 104, 836–847 (2000). Article CAS Google Scholar * Van
Stokkum, I. H. M., Larsen, D. S. & Van Grondelle, R. Global and target analysis of time-resolved spectra. _Biochim. Biophys. Acta Bioenerg._ https://doi.org/10.1016/j.bbabio.2004.04.011
(2004). * Holzwarth, A. R. Data Analysis of Time-Resolved Measurements. _Biophys. Tech. Photosynth._ https://doi.org/10.1007/0-306-47960-5_5 (2006). * Malavath, T., Caspy, I., Netzer-El, S.
Y., Klaiman, D. & Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. _Biochim. Biophys. Acta Bioenerg._ 1859, 645–654 (2018). Article
CAS PubMed Google Scholar * Nakane, T., Kimanius, D., Lindahl, E. & Scheres, S. H. W. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in
RELION. _Elife_. https://doi.org/10.7554/eLife.36861 (2018). * Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. _J. Struct. Biol._ 180,
519–530 (2012). Article CAS PubMed PubMed Central Google Scholar * Toporik, H., Li, J., Williams, D., Chiu, P. L. & Mazor, Y. The structure of the stress-induced photosystem I–IsiA
antenna supercomplex. _Nat. Struct. Mol. Biol._ 26, 443–449 (2019). Article CAS PubMed Google Scholar * Mazor, Y., Borovikova, A. & Nelson, N. The structure of plant photosystem I
super-complex at 2.8 A resolution. _Elife_ 4, e07433 (2015). Article PubMed PubMed Central Google Scholar * Qin, X., Suga, M., Kuang, T. & Shen, J.-R. Photosynthesis. Structural
basis for energy transfer pathways in the plant PSI-LHCI supercomplex. _Science_ 348, 989–995 (2015). Article ADS CAS PubMed Google Scholar * Mazor, Y., Borovikova, A., Caspy, I. &
Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 A resolution. _Nat. Plants_ 3, 17014 (2017). Article CAS PubMed Google Scholar * Brody, S. S. New excited state of
chlorophyll. _Science_. https://doi.org/10.1126/science.128.3328.838 (1958). * Gobets, B., Van Stokkum, I. H. M. M., Van Mourik, F., Dekker, J. P. & Van Grondelle, R. Excitation
wavelength dependence of the fluorescence kinetics in Photosystem I particles from Synechocystis PCC 6803 and Synechococcus elongatus. _Biophys. J._ 85, 3883–3898 (2003). Article CAS
PubMed PubMed Central Google Scholar * Brecht, M., Studier, H., Elli, A. F., Jelezko, F. & Bittl, R. Assignment of red antenna states in photosystem I from Thermosynechoccocus
elongatus by single-molecule spectroscopy. _Biochemistry_ 46, 799–806 (2007). Article CAS PubMed Google Scholar * Yin, S. et al. Assignment of the Qy, absorption spectrum of
photosystem-i from thermosynechococcus elongatus based on CAM-B3LYP calculations at the PW91-optimized protein structure. _J. Phys. Chem. B_ 111, 9923–9930 (2007). Article CAS PubMed
Google Scholar * Akita, F. et al. Structure of a cyanobacterial photosystem I surrounded by octadecameric IsiA antenna proteins. _Commun. Biol_. 3, 232 (2020). Article PubMed PubMed
Central CAS Google Scholar * Gisriel, C. et al. The structure of Photosystem I acclimated to far-red light illuminates an ecologically important acclimation process in photosynthesis.
_Sci. Adv._ 6, eaay6415 (2020). Article ADS CAS PubMed PubMed Central Google Scholar * Chen, M. et al. Distinct structural modulation of photosystem I and lipid environment stabilizes
its tetrameric assembly. _Nat. Plants_ 6, 314–320 (2020). Article CAS PubMed Google Scholar * Zheng, L. et al. Structural and functional insights into the tetrameric photosystem I from
heterocyst-forming cyanobacteria. _Nat. Plants_. https://doi.org/10.1038/s41477-019-0525-6 (2019). * Cao, P. et al. Structural basis for energy and electron transfer of the photosystem
I–IsiA–flavodoxin supercomplex. _Nat. Plants_ 6, 167–176 (2020). Article CAS PubMed Google Scholar * Li, M., Semchonok, D. A., Boekema, E. J. & Bruce, B. D. Characterization and
evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp TS-821. _Plant Cell_ 26, 1230–1245 (2014). Article CAS PubMed PubMed Central Google
Scholar * Lee, Y., Gorka, M., Golbeck, J. H. & Anna, J. M. Ultrafast energy transfer involving the red chlorophylls of cyanobacterial photosystem i probed through two-dimensional
electronic spectroscopy. _J. Am. Chem. Soc_. https://doi.org/10.1021/jacs.8b04593 (2018). * Wollman, F. A., Minai, L. & Nechushtai, R. The biogenesis and assembly of photosynthetic
proteins in thylakoid membranes. _Biochim. Biophys. Acta Bioenerg._ https://doi.org/10.1016/S0005-2728(99)00043-2 (1999). * Smart, L. B. & McIntosh, L. Expression of photosynthesis genes
in the cyanobacterium Synechocystis sp. PCC 6803:psaA-psaB and psbA transcripts accumulate in dark-grown cells. _Plant Mol. Biol_. https://doi.org/10.1007/BF00037136 (1991). * Zhu, Y.,
Liberton, M. & Pakrasi, H. B. A novel redoxin in the thylakoid membrane regulates the titer of photosystem I. _J. Biol. Chem_. https://doi.org/10.1074/jbc.M116.721175 (2016). *
Karapetyan, N. V., Dorra, D., Schweitzer, G., Bezsmertnaya, I. N. & Holzwarth, A. R. Fluorescence spectroscopy of the longwave chlorophylls in trimeric and monomeric photosystem I core
complexes from the cyanobacterium Spirulina platensis. _Biochemistry_. https://doi.org/10.1021/bi970386z (1997). * Karapetyan, N. V., Shubin, V. V. & Strasser, R. J. Energy exchange
between the chlorophyll antennae of monomeric subunits within the Photosystem I trimeric complex of the cyanobacterium Spirulina. _Photosynth. Res_. https://doi.org/10.1023/A:1006385002635
(1999). * Pishchalnikov, R. Y., Shubin, V. V. & Razjivin, A. P. Spectral differences between monomers and trimers of photosystem I depend on the interaction between peripheral
chlorophylls of neighboring monomers in trimer. _Phys. Wave Phenom_. https://doi.org/10.3103/S1541308X17030050 (2017). * Arnon, D. I. Copper enzymes in isolated chloroplasts.
polyphenoloxidase in beta vulgaris. _Plant Physiol_. https://doi.org/10.1104/pp.24.1.1 (1949). * Mastronarde, D. N. Automated electron microscope tomography using robust prediction of
specimen movements. _J. Struct. Biol_. https://doi.org/10.1016/j.jsb.2005.07.007 (2005). * Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution
single-particle cryo-EM. _Nat. Methods_. https://doi.org/10.1038/nmeth.2472 (2013). * Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron
microscopy. _Nat. Methods_ 14, 331 (2017). Article CAS PubMed PubMed Central Google Scholar * Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from
electron micrographs. _J. Struct. Biol._ 192, 216–221 (2015). Article PubMed PubMed Central Google Scholar * Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure
determination in RELION-3. _Elife_. https://doi.org/10.7554/eLife.42166 (2018). * Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in
cryo-EM single-particle analysis. _IUCrJ_. https://doi.org/10.1107/S205225251801463X (2019). * Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting.
_Structure_. https://doi.org/10.1016/j.str.2011.12.014 (2012). * Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. _Nat. Methods_.
https://doi.org/10.1038/nmeth.2727 (2014). * Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. _Acta
Crystallogr. Sect. D Struct. Biol_. https://doi.org/10.1107/S2059798319011471 (2019). * Cao, P. et al. Structural basis for energy and electron transfer of the photosystem I–IsiA–flavodoxin
supercomplex. _Nat. Plants_. https://doi.org/10.1038/s41477-020-0593-7 (2020). * Afonine, P. V., Headd, J. J., Terwilliger, T. C. & Adams, P. D. New tool: phenix.real_space_refine.
_Comput. Crystallogr. Newsl._ 4, 43–44 (2013). Google Scholar * Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. _Acta Crystallogr.
Sect. D_ 66, 213–221 (2010). Article CAS Google Scholar * Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. _Nat. Methods_.
https://doi.org/10.1038/s41592-020-0731-1 (2020). * Schrödinger, L. The PyMOL molecular graphics system. _Version_ 1, 8 (2015). Google Scholar * Pettersen, E. F. et al. UCSF Chimera-a
visualization system for exploratory research and analysis. _J. Comput. Chem._ 25, 1605–1612 (2004). Article CAS PubMed Google Scholar * Schlodder, E., Hussels, M., Çetin, M.,
Karapetyan, N. V. & Brecht, M. Fluorescence of the various red antenna states in photosystem i complexes from cyanobacteria is affected differently by the redox state of P700. _Biochim.
Biophys. Acta Bioenerg._ 1807, 1423–1431 (2011). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We would like to thank R. Blankenship for critical reading of the
manuscript. We would like to acknowledge the use of the Titan Krios at the Erying Materials Center at Arizona State University and the funding of this instrument by NSF MRI 1531991. This
study is funded by a startup grant from Arizona State University (to Y.M). The work performed at Kansas State University was supported by the Chemical Sciences, Geosciences and Biosciences
Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Grant No. DE-SC0006678 to R.J.). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of Molecular
Sciences, Arizona State University, Tempe, AZ, 85287-1604, USA Hila Toporik, Zachary Dobson, Reece Riddle, Su Lin & Yuval Mazor * Biodesign Center for Applied Structural Discovery,
Arizona State University, Tempe, AZ, 85287, USA Hila Toporik, Zachary Dobson, Reece Riddle, Su Lin & Yuval Mazor * Department of Chemistry, Kansas State University, Manhattan, KS, 66506,
USA Anton Khmelnitskiy & Ryszard Jankowiak * John M. Cowley Center for High Resolution Electron Microscopy, Arizona State University, Tempe, AZ, 85287, USA Dewight Williams * Center for
Innovations in Medicine at the Biodesign Institute, Arizona State University, Tempe, AZ, 85287, USA Su Lin * Department of Physics, Kansas State University, Manhattan, KS, 66506, USA
Ryszard Jankowiak Authors * Hila Toporik View author publications You can also search for this author inPubMed Google Scholar * Anton Khmelnitskiy View author publications You can also
search for this author inPubMed Google Scholar * Zachary Dobson View author publications You can also search for this author inPubMed Google Scholar * Reece Riddle View author publications
You can also search for this author inPubMed Google Scholar * Dewight Williams View author publications You can also search for this author inPubMed Google Scholar * Su Lin View author
publications You can also search for this author inPubMed Google Scholar * Ryszard Jankowiak View author publications You can also search for this author inPubMed Google Scholar * Yuval
Mazor View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Y.M., H.T., R.J., and A.K. performed experiments, analyzed data, and wrote the
manuscript. Z.D., R.R., S.L., and D.W. performed experiments. CORRESPONDING AUTHOR Correspondence to Yuval Mazor. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Communications_ thanks Mei Li, Fei Sun, and the other, anonymous reviewer(s) for their contribution to the peer review of
this work. Peer review reports are available. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTIONS OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY MOVIE 1 SUPPLEMENTARY MOVIE 2 SUPPLEMENTARY MOVIE 3
SUPPLEMENTARY MOVIE 4 SUPPLEMENTARY MOVIE 5 SUPPLEMENTARY MOVIE 6 REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative
Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the
original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in
the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended
use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Toporik, H., Khmelnitskiy, A., Dobson, Z. _et al._ The structure of a red-shifted
photosystem I reveals a red site in the core antenna. _Nat Commun_ 11, 5279 (2020). https://doi.org/10.1038/s41467-020-18884-w Download citation * Received: 18 May 2020 * Accepted: 11
September 2020 * Published: 19 October 2020 * DOI: https://doi.org/10.1038/s41467-020-18884-w SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative