Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

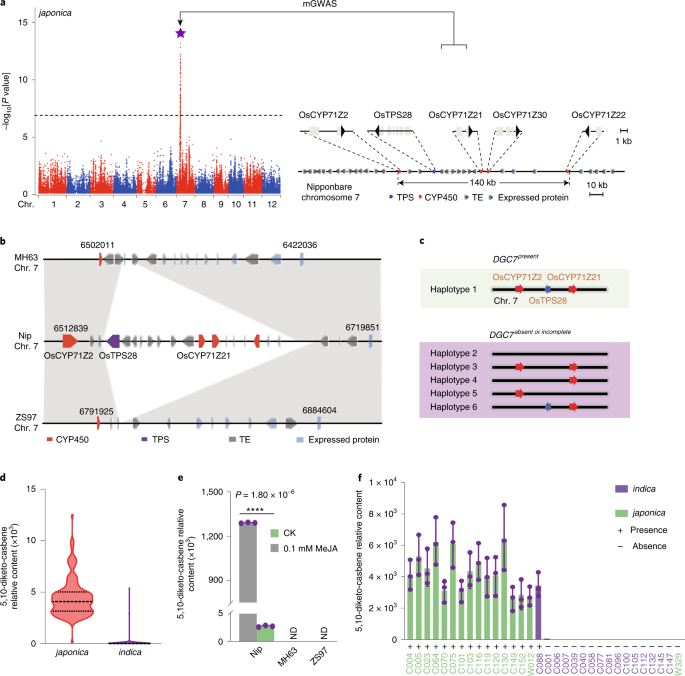
Diterpenoids are the major group of antimicrobial phytoalexins in rice1,2. Here, we report the discovery of a rice diterpenoid gene cluster on chromosome 7 (DGC7) encoding the entire
biosynthetic pathway to 5,10-diketo-casbene, a member of the monocyclic casbene-derived diterpenoids. We revealed that DGC7 is regulated directly by JMJ705 through methyl jasmonate-mediated
epigenetic control3. Functional characterization of pathway genes revealed OsCYP71Z21 to encode a casbene C10 oxidase, sought after for the biosynthesis of an array of medicinally important
diterpenoids. We further show that DGC7 arose relatively recently in the Oryza genus, and that it was partly formed in Oryza rufipogon and positively selected for in japonica during
domestication. Casbene-synthesizing enzymes that are functionally equivalent to OsTPS28 are present in several species of Euphorbiaceae but gene tree analysis shows that these and other
casbene-modifying enzymes have evolved independently. As such, combining casbene-modifying enzymes from these different families of plants may prove effective in producing a diverse array of
bioactive diterpenoid natural products.
The sequences data of 424 O. sativa accessions are available in the National Center for Biotechnology Information (NCBI) under the BioProject PRJNA171289 (ref. 17). The single nucleotide
polymorphisms information of 424 O. sativa accessions is available in RiceVarMap (http://ricevarmap.ncpgr.cn/v1). The pangenome data were acquired from the pangenome dataset
(https://figshare.com/collections/Novel_sequences_structural_variations_and_gene_presence_variations_of_Asian_cultivated_rice/3876022/1 and http://cgm.sjtu.edu.cn/3kricedb/)18,19,24.
Thirteen accessions of O. rufipogon were selected from 446 diverse O. rufipogon accessions from Asia and Oceania, and represented all the major genetically distinct clusters in O. rufipogon;
the other ten wild rice are from EnsemblPlants (http://plants.ensembl.org/index.html) and National Genomics Data Center (https://bigd.big.ac.cn/search?dbId=gwh&q=Oryza), including O.
barthii (AA), O. glumipatula (AA), O. glaberrima (AA), O. meridionalis (AA), O. longistaminata (AA), O. nivara (AA), O. brachyantha (FF), O. punctata (BB) and O. brachyantha (GG)24. Genes
reported in the study are deposited in the NCBI. The genes can be found in GenBank or Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml)
under the following accession numbers: OsTPS28, MN833254; OsCYP71Z21, LOC_Os07g11870; OsCYP71Z2, LOC_Os07g11739; OsCYP71Z22, LOC_Os07g11970; OsCYP71Z30, LOC_Os07g11890. Source data are
provided with this paper.
A Correction to this paper has been published: https://doi.org/10.1038/s41477-020-00838-1.
Schmelz, E. A. et al. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 79, 659–678 (2014).
We thank J. D. Keasling, G. P. Lomonossoff and Z. Zhao for their advice and their gift of the expression vectors and strains. We also thank D. R. Nelson, University of Tennessee, for help in
naming the OsCYP71Z30. This work was supported by the National Science Fund for Distinguished Young Scholars (grant no. 31625021), the State Key Programme of National Natural Science
Foundation of China (grant no. 31530052) and the Hainan University Startup Fund KYQD(ZR)1866 to J.L.
National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
Chuansong Zhan, Long Lei, Zixin Liu, Shen Zhou, Chenkun Yang, Xitong Zhu, Hao Guo, Feng Zhang, Meng Peng, Meng Zhang, Yufei Li, Zixin Yang, Yangyang Sun, Yuheng Shi, Kang Li, Shuangqian
Shen, Xuyang Wang, Jiawen Shao, Xinyu Jing, Zixuan Wang, Lianghuan Qu, Xianqing Liu, Ling-Ling Chen & Meng Yuan
Centre for Novel Agricultural Products, Department of Biology, University of York, York, UK
Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma, Japan
Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany
J.L. designed the research. J.L., L.-L.C., L.Q., M.Y. and X.L. supervised this study. C.Z., L. Lei, S.Z., Z.L., F.Z., M.Z., Y. Sun, Y. Shi, K.L., T.C., M.H., I.G., Z.Y. and T.T. participated
in the material preparation. C.Z., C.Y., Yi Li, X.W. and J.S. carried out the metabolite analyses. C.Z., Z.L., S.Z., C.Y., X.Z., H.G., M.P., M.Z., Yufei Li, Z.Y., L. Liu, S.S., J.S., X.J.,
Yi Li, T.T. and Z.W. performed the data analyses. C.Z., L. Lei, Z.L., S.Z. and C.Y. performed most of the experiments. J.L., C.Z., I.G. and A.R.F. wrote the manuscript.
Peer review information Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The core collection of 424 cultivated rice accessions in this study has a wide geographic distribution. Colour dots indicate different subspecies/type of cultivated rice.
a, Gas chromatography of the reaction products of OsTPS28 with GGDP. GGDP, geranylgeranyl diphosphate. Casbene and neocembrene reference compounds were purified from infiltrated N.
benthamiana leaves by the method described previously26. Compound 1, casbene; Compound 2, neocembrene. b, Gas chromatography of in vitro enzyme assays showing the 10-keto-casbene C5 oxidase
activity of yeast-expressed CYP71Z2 in the present of NADPH. Microsomes prepared from yeast containing PESC-URA empty vector were used as a negative control. 10-keto-casbene reference
compound was purified from rice leaves by the method described previously13,14. Compound 3, 10-keto- casbene; Compound 4, 5,10-diketo-casbene. c, Gas chromatography of the extracts prepared
from the leaves of N. benthamiana infiltrated with OsTPS28 overexpressing vector.
a, Mass spectrum and structure of the product in N. benthamiana leaves simultaneously overexpressing OsTPS28, OsCYP71Z2 and OsCYP71Z21. b, Mass spectrum of 5,10-diketo-casbene reference.
LC-MS, liquid chromatography–mass spectrometry. c, 1H NMR (left) and 13C NMR (right) results of 5,10-diketo-casbene.
The genes from DGC7 are indicated in bold. The transcript abundances of indicated genes in different organs at different stages were shown: expression levels of OsTPS28, OsCYP17Z2 and
OsCYP71Z21 is correlated at different developmental stages. The numerical values for blue-to-red gradient represent normalized expression levels from quantitative real-time PCR analysis.
The casbene-type diterpene biosynthetic pathways in rice and castor. Chr.7, chromosome 7; GGDP, geranylgeranyl diphosphate.
Anyone you share the following link with will be able to read this content: