A pharmacogenetic interaction analysis of bevacizumab with paclitaxel in advanced breast cancer patients
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT To investigate pharmacogenetic interactions among _VEGF-A_, _VEGFR-2_, _IL-8_, _HIF-1α_, _EPAS-1_, and _TSP-1_ SNPs and their role on progression-free survival (PFS) in metastatic
breast cancer (MBC) patients treated with bevacizumab plus first-line paclitaxel or with paclitaxel alone. Analyses were performed on germline DNA, and SNPs were investigated by real-time
PCR technique. The multifactor dimensionality reduction (MDR) methodology was applied to investigate the interaction between SNPs. The present study was an explorative, ambidirectional
cohort study: 307 patients from 11 Oncology Units were evaluated retrospectively from 2009 to 2016, then followed prospectively (NCT01935102). Two hundred and fifteen patients were treated
with paclitaxel and bevacizumab, whereas 92 patients with paclitaxel alone. In the bevacizumab plus paclitaxel group, the MDR software provided two pharmacogenetic interaction profiles
consisting of the combination between specific _VEGF-A_ rs833061 and _VEGFR-2_ rs1870377 genotypes. Median PFS for favorable genetic profile was 16.8 vs. the 10.6 months of unfavorable
genetic profile (_p_ = 0.0011). Cox proportional hazards model showed an adjusted hazard ratio of 0.64 (95% CI, 0.5–0.9; _p_ = 0.004). Median OS for the favorable genetic profile was 39.6
vs. 28 months of unfavorable genetic profile (_p_ = 0.0103). Cox proportional hazards model revealed an adjusted hazard ratio of 0.71 (95% CI, 0.5–1.01; _p_ = 0.058). In the 92 patients
treated with paclitaxel alone, the results showed no effect of the favorable genetic profile, as compared to the unfavorable genetic profile, either on the PFS (_p_ = 0.509) and on the OS
(_p_ = 0.732). The pharmacogenetic statistical interaction between _VEGF-A_ rs833061 and _VEGFR-2_ rs1870377 genotypes may identify a population of bevacizumab-treated patients with a better
PFS. SIMILAR CONTENT BEING VIEWED BY OTHERS AN INTEGRAL GENOMIC SIGNATURE APPROACH FOR TAILORED CANCER THERAPY USING GENOME-WIDE SEQUENCING DATA Article Open access 26 May 2022 SYSTEMATIC
PAN-CANCER ANALYSIS OF MUTATION–TREATMENT INTERACTIONS USING LARGE REAL-WORLD CLINICOGENOMICS DATA Article 30 June 2022 LESSONS LEARNED FROM A CANDIDATE GENE STUDY INVESTIGATING AROMATASE
INHIBITOR TREATMENT OUTCOME IN BREAST CANCER Article Open access 19 February 2025 INTRODUCTION The treatment of metastatic breast cancer (MBC) patients with hormone-receptors positive (HR+)
and human epidermal receptor 2 negative (HER2−) tumors is dramatically changed over the years. In this setting, cyclin-dependent kinase 4/6 inhibitors (CDK4/6i), such as palbociclib,
ribociclib and abemaciclib, in combination with aromatase inhibitors or fulvestrant represent today the first and later lines of therapy1. However, chemotherapy-based treatment is still a
therapeutic choice when hormone resistance occurs, in triple-negative tumor or in case of visceral crisis2,3. In this scenario, the humanized monoclonal antibody bevacizumab, in combination
with paclitaxel, is a treatment option compared to chemotherapy alone4. Although a significant improvement in progression-free survival (PFS) was observed from three comparative studies, the
US Food and Drug Administration (FDA), but not the European Medicines Agency (EMA), revoked the initial approval of bevacizumab for the first-line treatment of MBC patients because of the
lack of benefit in terms of overall survival (OS). However, it has been theorized that when a long survival post first-line progression is expected after a first-line chemotherapy, such as
in breast cancer, the lack of an apparent benefit in OS could not mean a lack of improvement in OS for the first line of treatment4,5,6,7,8. Different strategies have been investigated to
find possible predictive biomarkers and select those patients with the best chance of response to bevacizumab. Indeed, the PFS improvement due to bevacizumab was identical for magnitude in
all subgroups of patients with different clinical and pathological characteristics9, and therefore new selective biomarkers should be needed to identify those patients who can have a major
advantage in terms of outcome. Despite many attempts have been done, no validated biomarkers are currently available in the clinical practice and the prospective MERiDiAN trial failed to
demonstrate a possible role of VEGF-A baseline in predicting the response to bevacizumab in breast cancer patients10,11,12,13,14,15. Germline and somatic polymorphisms of genes involved in
the angiogenic pathways have also been widely investigated in this research area to predict bevacizumab outcome, with contrasting results12,16,17,18. Due to the retrospective nature of these
studies and to their inconclusive results, the role of single nucleotide polymorphisms (SNPs) as predictive markers remains to define19. Therefore, the current approach of correlating the
bevacizumab response to a single SNP may be replaced by a genetic analysis of the interaction between SNPs, defined as non-linear interaction or epistasis. Moore and colleagues have
established and validated a methodology, called multifactor dimensionality reduction (MDR) analysis, to identify a genetic profile with the ability to predict the drug response20. To test
this hypothesis, we conducted a retrospective study on 113 MBC patients to assess the ability of MDR methodology to identify a favorable pharmacogenetic profile associated to PFS in patients
treated with bevacizumab, combined with first-line paclitaxel. The MDR analysis provided two pharmacogenetic interaction profiles consisting of the combination between specific _VEGFR-2_
rs11133360 and _IL-8_ rs4073 genotypes. The median PFS was 14.1 months (95% CI, 11.4–16.8) and 10.2 months (95% CI, 8.8–11.5) for the favorable and the unfavorable genetic profile,
respectively (HR = 0.44; 95% CI, 0.29–0.66; _p_ < 0.0001)21. Based on these encouraging results, our study was planned to evaluate the effects of the combination of paclitaxel with
bevacizumab on patients harboring other different genetic profiles, exploring the possibility to predict the best favorable profile in terms of PFS, our primary endpoint. The second step was
to test if the eventual seen PFS advantage could be maintained also in terms of OS (our secondary endpoint) in these patients even after the end of the administration of bevacizumab
combined therapy. The analysis was extended to a group of patients treated without bevacizumab, during the same period of time, with the purpose of having a control group. RESULTS PATIENTS
Two-hundred and fifteen patients treated with bevacizumab in combination with paclitaxel and 92 patients treated with first-line chemotherapy without bevacizumab, entered the present MDR
analysis. In the bevacizumab plus paclitaxel group, the median number of cycles administered were 7 (range 4–18) and maintenance with bevacizumab alone was continued in 152 patients (70.7%).
All the 215 patients were evaluated for the response. 21 (10%) and 126 (58%) experienced a complete and a partial response, respectively; 52 patients (24%) reported a stable disease (SD)
and in 16 patients (8%) a progression was observed. None of the analyzed polymorphisms was associated with the response rate (data not shown). When the present analysis was performed, 215
out of 215 patients (100%) progressed and 170 out of 215 patients (79%) died from the metastatic disease. No patients died of cancer-unrelated causes. The median PFS and median OS were 11.8
months (95% CI, 10.9–12.7 months) and 30.7 months (95% CI, 26–35.5 months), respectively. Data were censored after 70 months. The main characteristics of the patients are reported in Table
1. While the differences observed between groups may be due to the retrospective nature of the study, the characteristics between favorable and unfavorable group were superimposable.
ASSOCIATION OF CLINICAL AND PATHOLOGICAL CHARACTERISTICS WITH PFS AND OS In Table 2 are reported the associations of clinical and pathological characteristics with both PFS and OS in the 215
patients treated with paclitaxel and bevacizumab. Hormonal-receptor status confirmed its role in determining the prognosis of this group of patients. Interestingly, in the group of patients
who continued bevacizumab, over the patients who interrupted it at the end of chemotherapy without evidence of a disease progression, both a greater PFS and OS was observed. In Table 3 are
reported the associations of clinical and pathological characteristics with PFS in the group of patients (_n_ = 92 that received paclitaxel without bevacizumab). The Cox proportional hazards
analysis was applied to evaluate the association between each single polymorphism with both PFS and OS. The analysis did not reveal significant positive association between each SNP with
PFS (Table 4). No significant associations were observed with OS (data not shown). MDR ANALYSIS The MDR analysis revealed a genetic interaction profile, consisting of the combination between
specific _VEGF-A_ rs833061 and _VEGFR-2_ rs1870377 genotypes, significantly associated with PFS and OS benefit. Particularly, two pharmacogenetic profiles were identified in patients, as
reported in Table 5. The first one was associated with a greater PFS and OS benefit, whereas the second one with a lower PFS and OS after paclitaxel plus bevacizumab treatment. The
characteristics at baseline of patients treated with paclitaxel alone or paclitaxel + bevacizumab harboring the pharmacogenetic favorable and unfavorable profile are reported in Tables 6 and
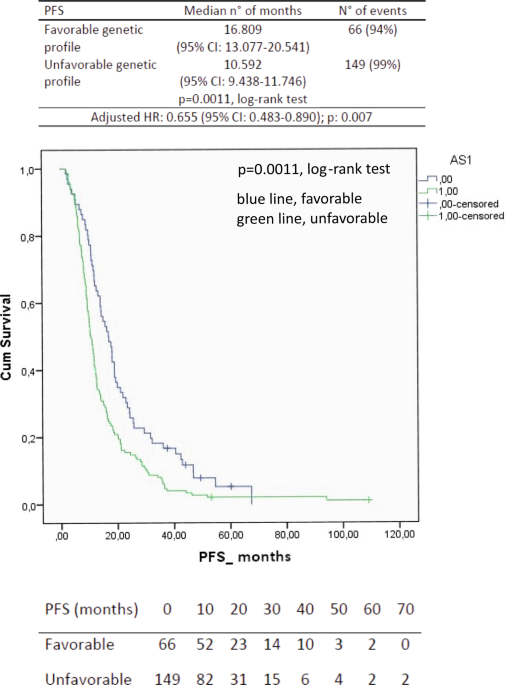
7, respectively. The median PFS for the favorable genetic profile was 16.8 months (95% CI, 13.1–20.5 months) vs. the 10.6 months of the unfavorable genetic profile (95% CI, 9.4–11.7 months;
_p_ = 0.0011, log-rank test; Fig. 1). The Cox proportional hazards model, which was performed to assess the adjusted hazard ratio for the PFS of the favorable genetic profile, showed a
value of 0.64 (95% CI, 0.5–0.9; _p_ = 0.004; Table 8). Furthermore, a formal test of interaction confirmed the predictive nature of the favorable profile in the bevacizumab + paclitaxel
group as reported in supplementary Table 1. Remarkably, the patients included in the favorable genetic profile also had the best probability of OS benefit, and the difference was significant
as compared to the OS of the unfavorable genetic profile (Fig. 2). The median OS for the favorable genetic profile was 39.6 months (95% CI, 30.2–40.1 months) vs. the 28 months of the
unfavorable genetic profile (95% CI, 24–32 months; _p_ = 0.0103, log-rank test; Fig. 2). The Cox proportional hazards model, including the same significant parameters described in Table 8,
revealed an adjusted hazard ratio for the OS of the favorable genetic profile of 0.71 (95% CI, 0.5–1.01; _p_ = 0.058), at the limit of significance. Of note, the probability of an estimated
1-year survival rate was 90.9% (95% CI, 90.4–91.4) in the favorable genetic profile and 80.5% (95% CI, 80.2–80.8) in the unfavorable genetic profile; the estimated 2-year survival was 75.7%
(95% CI, 75.2–76.2) and 57% (95% CI, 56–57.4), respectively. The estimated 3-year survival rate was 56.1% (95% Cl, 56.1–57.1) in the favorable genetic profile and 38.9% (95% Cl, 38.4–39.3)
in the unfavorable. The observed objective responses were 69.7% in the favorable genetic profile as compared with 69.1% in the unfavorable genetic profile. Also, the 92 MBC patients treated
with a first-line chemotherapy including paclitaxel without bevacizumab were investigated to test the impact of the two genetic profiles in both PFS and OS. The results revealed no effect of
the favorable genetic profile, as compared to the unfavorable genetic profile, either on the PFS (_p_ = 0.509, log-rank test; Supplementary Fig. 1a) or on the OS (_p_ = 0.732, log-rank
test; Supplementary Fig. 1b). DISCUSSION The current standard therapy of patients suffering of metastatic breast cancer with HR+ and HER2− disease, in first and later lines of treatment, is
represented by combinations of hormone and novel targeted therapies. The CDK4/6i palbociclib, ribociclib, and abemaciclib in combination with aromatase inhibitors or fulvestrant have
dramatically changed the treatment of this setting of patients1. Despite these treatments’ advances, the chemotherapy maintains its key role because almost all metastatic patients with HR+
and HER2− disease develop resistance over time to endocrine therapy. As well, chemotherapy represents the first choice of treatment in triple negative, _BRCA_ wild type and PD-L1 negative
disease3. In this _scenario_, bevacizumab in combination with paclitaxel can still represent an option but the lack of any advantage in terms of OS, led to a slow decline in its use in the
clinical practice during last years. Moreover, a recent meta-analysis has investigated, in head-to-head comparison, the role of endocrine treatment _versus_ chemotherapy in postmenopausal
setting with HR+ and HER2− metastatic disease, highlighting that bevacizumab in combination with paclitaxel was the only regimen that was significantly better than palbociclib plus letrozole
in terms of response rate22. Thus, the identification of pharmacodynamic biomarkers could better select patients with the best chance of bevacizumab response and clarify the role of this
antiangiogenic antibody in the management of MBC patients. The multifactor dimensionality reduction (MDR) methodology has been previously used to identify genetic polymorphisms interactions
profiles able in predicting drug response in metastatic colorectal cancer patients. In the study published by Pander and colleagues23, an interaction between VEGF+ 405G>C and TYMS-TSER
polymorphisms, instead of an individual polymorphism, seemed to predict the CAPOX-B (capecitabine, oxaliplatin, and bevacizumab combination) response in terms of PFS, suggesting a paradigm
shift from SNPs to a more complex interaction gene analysis able to predict response to antitumor agents. The current study was planned to evaluate the effects of the combination of
paclitaxel with bevacizumab _versus_ paclitaxel alone on MBC patients harboring different genetic profiles, exploring the possibility to predict the best favorable profile in terms of PFS.
The second step was to test if the eventual seen PFS advantage could be maintained also in terms of OS in these patients. The previous study on _VEGFR-2_ and _IL-8_ genetic interaction
analysis21, the favorable profile in terms of PFS was not predictive of OS benefit. In the present study, the seen advantage in PFS was indeed confirmed in OS (an adjusted hazard ratio of
0.71) but with a _p_ = 0.058, a value very close to a statistically significance, but not significant. However, the reported data, although statistically negative, seem to suggest that the
favorable profile in terms of PFS may probably be maintained also in terms of OS and undoubtedly merits further investigations in a validation prospective study. Indeed, evaluation and
confirmation of these findings in an independent cohort is critical because of the exploratory nature of our ambidirectional trial. The analyses conducted with the MDR methodology in this
unselected population of MBC patients revealed more than a genetic interaction profile, consisting of the combination between specific genotypes, but, due to nature of the MDR methodology,
we investigated the genetic profile with a benefit in terms of both PFS and OS. The analysis conducted revealed a genetic interaction profile, consisting of the combination between specific
genotypes of _VEGF_ rs833061 and _VEGFR-2_ rs1870377. Particularly, two genetic profiles were identified in patients, as reported in Table 5. The first one was associated with a greater both
PFS and OS benefit compared to the second one. However, this model considered all the candidates and allows for any and all combination of SNPs to correlate with outcome. Thus, there are
other significant or borderline permutations. Indeed, we have also included, as an example in the supplementary data (Supplementary Table 2 and Supplementary Fig. 2), another interesting
genetic profile with a significant advantage in term of PFS but without any advantage in OS (not even a tendency). Therefore, in our study we demonstrated, through the MDR methodology, a
statistical interaction between _VEGF-A_ and _VEGFR-2_ gene SNPs that potentially relates to bevacizumab efficacy on both PFS and OS. The two genes, and, consequently, the two proteins
belong to the same signaling pathway, and it has been clearly demonstrated that VEGF-A stimulates VEGFR-2 phosphorylation and tumor angiogenesis24. Based on these premises, it is conceivable
to hypothesize that, in patients carrying the favorable genetic profile, the tumor angiogenesis is successfully inhibited in the presence of bevacizumab. The pharmacological inhibition of
the angiogenic process by bevacizumab could be effective because of the physiological (not increased) production of VEGF-A due to the presence of _VEGF-A_ rs833061 CC genotype or C allele.
Indeed, for this SNP _VEGF-A_ rs833061 C>T it has been described an increased promoter activity due to the T allele25 that may explain an eventual resistance to the treatment. Moreover,
the _VEGFR-2_ rs1870377 is a nonsynonymous SNP substituting glycine with histidine (Q472H) located in the extracellular ligand binding region of the receptor, potentially impacting VEGFR-2
degradation26. The _VEGFR-2_ rs1870377 TT genotype or T allele present in the favorable profile synthetize a receptor not modified in its structure, suggesting that it is not abnormally
activated or degraded. Therefore, it might be plausible that the genetic background characterized by a physiological activation of the VEGF-A pathway may be responsible, in part, for the
positive effect of bevacizumab maintenance therapy in these MBC patients. In contrast, in patients with an unfavorable genetic profile, the microenvironment conditions due to the different
genotype combinations may result in an increase of the VEGF-A production and/or the presence of an altered VEGFR-2 on tumor endothelial cells which may be capable to proliferate, migrate or
survive because the VEGF action is not completely blocked by bevacizumab. The absence of any advantage in terms of efficacy in the patients treated with chemotherapy without bevacizumab
could suggest a possible predictive role of the favorable genetic profile for bevacizumab response, but the exploratory nature of this ambidirectional study may limit this hypothesis.
However, the main findings of our analyses support the conclusion that a genetic profile may identify a group of patients with longer PFS and OS, predicting the response to bevacizumab in
combination with paclitaxel. The MDR approach is a major reason for differences between our trial and other studies on bevacizumab biomarkers such as E210016. There are additional aspects
between the E2100 US patients and the Italian population of our study that may account for different results. First of all, Italian patients were of Caucasian origin and no patients of
African origin were represented. Secondly, although the frequencies of our studied _VEGF-A_ and _VEGFR-2_ SNPs were superimposable with the ones of the Caucasian patients published in the
article by Schneider and colleagues16, there was an exception regarding the VEGFR-2 889A/G (rs2071559). In that case, the frequency of the minor allele A in our population was 0.49 whereas
in the E2100 study was 0.09. New pharmacogenetic favorable biomarkers of bevacizumab-combined therapies could be retrieved from a genetic analysis of the interaction among SNPs rather than
from the examination of a single SNP of a single gene. Surely, a multigene-risk biomarkers may be more beneficial from a comprehensive agnostic approach using genome-wide association studies
(GWAS) rather than a candidate gene approach as the one that we have used in our study. However, some challenges have been faced when scientists tried the scaling of MDR to big data, as the
one from GWAS, such as the necessity to filter the data prior to MDR analysis27, also using biological knowledge through tools such as BioFilter28. Moreover, our work can definitively be
strengthened by the biological characterization of the VEGF expression in the pre-treatment tissue. Indeed, since rs833061 is located in the promoter region of VEGF-A, the difference in
expression levels of VEGF-A in tumors of patients harboring the favorable vs. unfavorable profile could be an important strategy to confirm our statistical findings. In conclusion, the MDR
methodology could be successfully used as witnessed by the experience in this unselected MBC patients where the investigation of an interaction between _VEGF-A rs833061 and VEGFR-2
rs1870377_ gene polymorphisms resulted in the identification of a genetic profile associated with a longer PFS. METHODS STUDY POPULATION This is an explorative, ambidirectional cohort study,
meaning that eligible patients were enrolled and evaluated retrospectively from January 2009 until September 2016 and then followed prospectively. The oncology units, all located in the
north or center of Italy, were selected based on their clinical experience in the use of the combination of paclitaxel and bevacizumab as first-line therapy in histologically confirmed
HER-2-negative MBC patients. Two-hundred fifteen patients from 11 Italian divisions of Medical Oncology, with histologically confirmed HER2-negative MBC, were treated with a first-line
therapy including bevacizumab 10 mg/m2 i.v. on days 1 and 15 combined with first-line paclitaxel 90 mg/m2 i.v. on days 1, 8, and 15, every 4 weeks, and they were enrolled for the present
pharmacogenetic study. Ninety-two MBC patients treated with a first-line chemotherapy including paclitaxel without bevacizumab, during the same period, were also included into the study as a
control group. The patients enrolled in the previously published study21 have been also included in the present analysis. Basal and pathological characteristics recorded from both groups
were the following: age (≤ or >65 years); Eastern Cooperative Oncology Group (ECOG) performance status (0 or 1–2); hormonal-receptor status (positive or negative); previous adjuvant
chemotherapy (none, anthracycline or anthracycline plus taxanes); previous hormonal therapy (adjuvant or metastatic); disease-free interval from the first diagnosis of breast cancer (≤ or
>12 months); extent of disease (≤ or >3 sites); location of disease (viscera or bone); disease evaluation (measurable or non-measurable). Patients with human epidermal growth factor
receptor type 2 (HER2)-positive, were excluded from the present study. The characteristics of the patients are summarized in Table 1. The treatment with chemotherapy was continued until
either disease progression occurred or unacceptable toxicities registered, or it was stopped for medical choice. The bevacizumab maintenance was continued, and hormone therapy added for both
groups when indicated, until disease progression or unacceptable toxicities occurred. Sites of metastatic disease were radiologically re-evaluated according to the RECIST criteria 1.1, in
patients with measurable disease, every 2 months. In patients without measurable lesions, progression of disease was defined when new lesions appeared or when existing lesions evolved.
Likewise, in the case of non-measurable lesions, deterioration of clinical condition not due to treatment toxicity, was defined as progression of disease. PFS was defined as the period from
the beginning of the treatment to the first observation of disease progression as above described, or death from any cause. OS was defined as the period from the beginning of the treatment
to death from any cause. All patients were assessed for response, PFS and OS. Each patient entering the study signed the informed consent. The disease assessment was conducted by the
investigators based on the approved protocol and all the oncology units followed the same assessment schedule and criteria for the prospective follow-up. The protocol was approved by ethic
committee of Azienda Ospedaliera-Universitaria Pisana (CESM-AOUP 3077/2010; clinicaltrials.gov identifier NCT01935102) for Pisa, Livorno, Lucca, Massa Carrara, Versilia, and Pontedera
Hospitals, and by the ethic committees of all participating centers. GENOTYPING ANALYSES Blood samples (3 ml) were collected in EDTA tubes and stored at −80 °C. Genes and polymorphisms,
involved in the angiogenesis pathways, were selected for the present analyses based on our previous study21. In the Table 9, the selected polymorphisms are reported. Germline DNA extraction
was performed using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA). Allelic discrimination of genes was performed using an ABI PRISM 7900 SDS (Applied Biosystems, Carlsbad, CA, USA)
and with validated TaqMan® SNP genotyping assays (Table 9; Applied Biosystems). PCR reactions were carried out according to the manufacturer’s protocol. Genotyping was not performed until an
adequate number of events (>80% on study population) was reported in terms of PFS. All the samples were analyzed twice to replicate the obtained genotype. STATISTICAL ANALYSIS The
investigators responsible for data analysis were blinded to which samples were from patients treated with paclitaxel alone and paclitaxel plus bevacizumab. The aim of the present study was
to identify a favorable genetic profile in terms of PFS in MBC patients treated with bevacizumab in association with paclitaxel. The corresponding OS in these patients remained a secondary
endpoint as well as response rate. All polymorphisms were analysed for deviation from the Hardy–Weinberg Equilibrium (HWE) by means of comparison between observed allelic distributions with
those expected from the HWE by on _χ_2 test (see Supplementary Tables 3 and 4). Any association between gene polymorphisms and response rate was analysed by the two-sided Fisher’s exact
test. The association between each individual polymorphism and the most relevant clinical-pathological characteristics with PFS and OS was tested using a Cox proportional hazards model. In
these analyses we used Bonferroni’s correction and the _p_ value <0.00357 (0.05/14 SNPs = 0.00357) was accepted as statistically significant. The multifactor dimensionality reduction
(MDR) methodology was applied (MDR software version 2.0 beta 6 on http://sourceforge.net/projects/mdr/, last access January 2021) to investigate the interaction between gene polymorphisms
and to identify favorable genetic profiles associated with the greater PFS in this population of patients. MDR was developed as a non-parametric and genetic model-free data mining strategy
for identifying combinations of SNPs that are predictive of a discrete clinical endpoint. MDR approach is a constructive induction algorithm that creates a new attribute by pooling genotypes
from multiple SNPs29,30. The difference both in PFS and OS between favorable genetic profiles and the unfavorable genetic profiles were assessed with the log-rank test and the Kaplan–Meier
method to evaluate survival curves. A Cox proportional hazards model, with the possible genetic profiles and the clinical and pathological patient’s characteristics individually related with
both the PFS and OS, was used to calculate the adjusted hazards ratio (HR) and the 95% confidence interval (95% CI). The Kaplan–Meier and Cox proportional hazards analyses were performed
using the SPSS version 17.0 (SPSS, Chicago, IL). For the genotype combination we used a statistical correction. Indeed, the _p_ value for the statistical significance was obtained using
1000-fold permutation testing (software available on https://sourceforge.net/projects/mdr/files/mdrpt/), and the significance was set for values less than 0.05. As an explorative study in
nature, no estimation of power and sample size was performed because of the absence of previous published data regarding the specific investigated genetic profiles and the administered
combination treatment. No data were excluded from the analysis. REPORTING SUMMARY Further information on research design is available in the Nature Research Reporting Summary linked to this
article. DATA AVAILABILITY The data that support the findings of this study are available from the corresponding author upon reasonable request. CODE AVAILABILITY The code for calculating
HRs is written in SPSS version 17.0 and the code for identifying favorable/unfavorable profiles is written in Multifactor Dimensionality Reduction software version 2.0 beta 6. Codes are
available upon request to M.S. REFERENCES * Schettini, F. et al. Overall survival of CDK4/6-inhibitor-based treatments in clinically relevant subgroups of metastatic breast cancer:
systematic review and meta-analysis. _J. Natl Cancer Inst._ 112, 1089–1097 (2020). Article CAS Google Scholar * AIOM. Linee guida Neoplasia della mammella.
https://www.aiom.it/wp-content/uploads/2020/10/2020_LG_AIOM_Neoplasie_Mammella_16022021.pdf (2020). * Manjunath, M. & Choudhary, B. Triple‑negative breast cancer: a run‑through of
features, classification and current therapies (Review). _Oncol. Lett._ 22, 512 (2021). Article CAS Google Scholar * Miller, K. et al. Paclitaxel plus bevacizumab versus paclitaxel alone
for metastatic breast cancer. _N. Engl. J. Med._ 357, 2666–2676 (2007). Article CAS Google Scholar * Rugo, H. S. & Diller, H. Inhibiting angiogenesis in breast cancer: the beginning
of the end or the end of the beginning? _J. Clin. Oncol._ 30, 898–901 (2012). Article Google Scholar * Miles, D. W. et al. Phase III study of bevacizumab plus docetaxel compared with
placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. _J. Clin. Oncol._ 28, 3239–3247 (2010). Article CAS
Google Scholar * Robert, N. J. et al. RIBBON-1: randomized, double-blind, placebo-controlled, phase III trial of chemotherapy with or without bevacizumab for first-line treatment of human
epidermal growth factor receptor 2-negative, locally recurrent or metastatic breast cancer. _J. Clin. Oncol._ 29, 1252–1260 (2011). Article CAS Google Scholar * Broglio, K. R. &
Berry, D. A. Detecting an overall survival benefit that is derived from progression-free survival. _J. Natl Cancer Inst._ 101, 1642–1649 (2009). Article Google Scholar * O’Shaughnessy, J.
et al. A meta-analysis of overall survival data from three randomized trials of bevacizumab (BV) and first-line chemotherapy as treatment for patients with metastatic breast cancer (MBC).
_J. Clin. Oncol._ 28, 1005 (2010). Article Google Scholar * Van Cutsem, E. et al. 803 ORAL analysis of blood plasma factors in the AVITA phase III randomized study of bevacizumab (bev)
with gemcitabine-erlotinib (GE) in patients (pts) with metastatic pancreatic cancer (mPC). _Eur. J. Cancer_ 47, S95–S96 (2011). Article Google Scholar * Van Cutsem, E. et al. Bevacizumab
in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. _J. Clin. Oncol._ 30, 2119–2127 (2012).
Article Google Scholar * Miles, D. W. et al. Biomarker results from the AVADO phase 3 trial of first-line bevacizumab plus docetaxel for HER2-negative metastatic breast cancer. _Br. J.
Cancer_ 108, 1052–1060 (2013). Article CAS Google Scholar * Jayson, G. C., Hicklin, D. J. & Ellis, L. M. Antiangiogenic therapy-evolving view based on clinical trial results. _Nat.
Rev. Clin. Oncol._ 9, 297–303 (2012). Article CAS Google Scholar * Hegde, P. S. et al. Predictive impact of circulating vascular endothelial growth factor in four phase III trials
evaluating bevacizumab. _Clin. Cancer Res._ 19, 929–937 (2013). Article CAS Google Scholar * Lambrechts, D., Lenz, H. J., De Haas, S., Carmeliet, P. & Scherer, S. J. Markers of
response for the antiangiogenic agent bevacizumab. _J. Clin. Oncol._ 31, 1219–1230 (2013). Article CAS Google Scholar * Schneider, B. P. et al. Association of vascular endothelial growth
factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG
2100. _J. Clin. Oncol._ 26, 4672–4678 (2008). Article CAS Google Scholar * Etienne-Grimaldi, M. C. et al. Prospective analysis of the impact of VEGF-A gene polymorphisms on the
pharmacodynamics of bevacizumab-based therapy in metastatic breast cancer patients. _Br. J. Clin. Pharm._ 71, 921–928 (2011). Article CAS Google Scholar * Loupakis, F. et al.
Retrospective exploratory analysis of VEGF polymorphisms in the prediction of benefit from first-line FOLFIRI plus bevacizumab in metastatic colorectal cancer. _BMC Cancer_ 11, 1–9 (2011). *
Bocci, G. & Loupakis, F. Bevacizumab pharmacogenetics in tumor treatment: still looking for the right pieces of the puzzle. _Pharmacogenomics_ 12, 1077–1080 (2011). * Pan, Q., Hu, T.
& Moore, J. H. Epistasis, complexity, and multifactor dimensionality reduction. _Methods Mol. Biol._ 1019, 465–477 (2013). Article CAS Google Scholar * Allegrini, G. et al.
Pharmacogenetic interaction analysis of VEGFR-2 and IL-8 polymorphisms in advanced breast cancer patients treated with paclitaxel and bevacizumab. _Pharmacogenomics_ 15, 1985–1999 (2014).
Article CAS Google Scholar * Giuliano, M. et al. Endocrine treatment versus chemotherapy in postmenopausal women with hormone receptor-positive, HER2-negative, metastatic breast cancer: a
systematic review and network meta-analysis. _Lancet Oncol._ 20, 1360–1369 (2019). Article CAS Google Scholar * Pander, J. et al. Pharmacogenetic interaction analysis for the efficacy of
systemic treatment in metastatic colorectal cancer. _Ann. Oncol._ 22, 1147–1153 (2011). Article CAS Google Scholar * Wang, X., Bove, A. M., Simone, G. & Ma, B. Molecular bases of
VEGFR-2-mediated physiological function and pathological role. _Front. Cell Dev. Biol_. 8, 1314 (2020). * Stevens, A., Soden, J., Brenchley, P. E., Ralph, S. & Ray, D. W. Haplotype
analysis of the polymorphic human vascular endothelial growth factor gene promoter. _Cancer Res_ 63, 812–816 (2003). CAS PubMed Google Scholar * Gal, J. et al. VEGF-Related germinal
polymorphisms may identify a subgroup of breast cancer patients with favorable outcome under bevacizumab-based therapy—a message from COMET, a French Unicancer Multicentric Study.
_Pharmaceuticals_ 13, 414 (2020). Article CAS Google Scholar * Sun, X. et al. Analysis pipeline for the epistasis search—statistical versus biological filtering. _Front. Genet_. 5, 106
(2014). * Pendergrass, S. A. et al. Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. _BioData Min_. 6, 1–20 (2013). * Hahn, L. W. &
Moore, J. H. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. _Silico Biol._ 4, 183–194 (2004). CAS Google Scholar * Moore, J. H. et al. A flexible
computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. _J. Theor. Biol._ 241, 252–261
(2006). Article Google Scholar Download references ACKNOWLEDGEMENTS The present work was supported by The Fondazione per le Attività di Ricerca in Oncologia (F.A.R.O.) of Pontedera, Pisa,
(Italy) and by the Italian Association for Cancer Research (AIRC) to G.B. The authors thank the nurse, laboratory, and administrative staff of all participating centers for their precious
help. We would like to give special thanks to the friends of “Deportivo Peccioli”, to the Municipality of Peccioli, Pisa, Italy and to the “Amici di Antonella Onlus” who supported the
project. AUTHOR INFORMATION Author notes * These authors contributed equally: Luigi Coltelli, Giacomo Allegrini, Paola Orlandi. AUTHORS AND AFFILIATIONS * Department of Oncology, Azienda USL
Toscana Nord Ovest, Pisa, Italy Luigi Coltelli, Giacomo Allegrini, Chiara Finale, Luna Chiara Masini, Giada Arrighi, Maria Teresa Barletta, Ermelinda De Maio, Samanta Cupini, Francesca
Orlandi, Damiana Francesca, Leonardo Barellini & Alessandro Cosimi * Division of Medical Oncology, Livorno and Pontedera Hospitals, Azienda USL Toscana Nord Ovest, Pisa, Italy Luigi
Coltelli, Giacomo Allegrini, Chiara Finale, Luna Chiara Masini, Giada Arrighi, Maria Teresa Barletta, Ermelinda De Maio, Samanta Cupini & Francesca Orlandi * Department of Clinical and
Experimental Medicine, University of Pisa, Pisa, Italy Paola Orlandi, Marta Banchi, Elisabetta Fini, Patrizia Guidi, Giada Frenzilli & Guido Bocci * Division of Medical Oncology II,
Azienda Ospedaliero-Universitaria Pisana, S. Chiara Hospital, Pisa, Italy Andrea Fontana, Barbara Salvadori, Giulia Lorenzini & Alfredo Falcone * Institute of Clinical Physiology,
Italian National Research Council – CNR, Pisa, Italy Marco Scalese * Division of Medical Oncology, Versilia Hospital, Azienda Usl Toscana Nord Ovest, Lido di Camaiore, Italy Sara Donati *
Division of Medical Oncology, San Luca Hospital, Azienda Usl Toscana Nord Ovest, Lucca, Italy Simona Giovannelli & Lucia Tanganelli * Division of Radiotherapy, Azienda
Ospedaliero-Universitaria Careggi, Firenze, Italy Lorenzo Livi & Icro Meattini * Division of Medical Oncology, Pescia and Pistoia Hospitals, Azienda Usl Toscana Centro, Pistoia, Italy
Ilaria Pazzagli & Marco Di Lieto * Division of Medical Oncology, Umberto I Salesi-Lancisi Hospital, Azienda Ospedaliero-Universitaria Umberto I, Ancona, Italy Mirco Pistelli * Division
of Medical Oncology, Marche Nord Hospital, Azienda Ospedaliera San Salvatore, Pesaro, Italy Virginia Casadei * Division of Medical Oncology, Santa Chiara Hospital, Azienda Provinciale per I
Servizi Sanitari, Trento, Italy Antonella Ferro * Division of Radiology, Pontedera Hospital, Azienda Usl Toscana Nord Ovest, Pisa, Italy Damiana Francesca * Breast Unit – Division of Breast
Surgery, Livorno Hospital, Azienda Usl Toscana Nord Ovest, Livorno, Italy Leonardo Barellini * Department of Translational Research and New Technology in Medicine and Surgery, University of
Pisa, Pisa, Italy Alfredo Falcone Authors * Luigi Coltelli View author publications You can also search for this author inPubMed Google Scholar * Giacomo Allegrini View author publications
You can also search for this author inPubMed Google Scholar * Paola Orlandi View author publications You can also search for this author inPubMed Google Scholar * Chiara Finale View author
publications You can also search for this author inPubMed Google Scholar * Andrea Fontana View author publications You can also search for this author inPubMed Google Scholar * Luna Chiara
Masini View author publications You can also search for this author inPubMed Google Scholar * Marco Scalese View author publications You can also search for this author inPubMed Google
Scholar * Giada Arrighi View author publications You can also search for this author inPubMed Google Scholar * Maria Teresa Barletta View author publications You can also search for this
author inPubMed Google Scholar * Ermelinda De Maio View author publications You can also search for this author inPubMed Google Scholar * Marta Banchi View author publications You can also
search for this author inPubMed Google Scholar * Elisabetta Fini View author publications You can also search for this author inPubMed Google Scholar * Patrizia Guidi View author
publications You can also search for this author inPubMed Google Scholar * Giada Frenzilli View author publications You can also search for this author inPubMed Google Scholar * Sara Donati
View author publications You can also search for this author inPubMed Google Scholar * Simona Giovannelli View author publications You can also search for this author inPubMed Google Scholar
* Lucia Tanganelli View author publications You can also search for this author inPubMed Google Scholar * Barbara Salvadori View author publications You can also search for this author
inPubMed Google Scholar * Lorenzo Livi View author publications You can also search for this author inPubMed Google Scholar * Icro Meattini View author publications You can also search for
this author inPubMed Google Scholar * Ilaria Pazzagli View author publications You can also search for this author inPubMed Google Scholar * Marco Di Lieto View author publications You can
also search for this author inPubMed Google Scholar * Mirco Pistelli View author publications You can also search for this author inPubMed Google Scholar * Virginia Casadei View author
publications You can also search for this author inPubMed Google Scholar * Antonella Ferro View author publications You can also search for this author inPubMed Google Scholar * Samanta
Cupini View author publications You can also search for this author inPubMed Google Scholar * Francesca Orlandi View author publications You can also search for this author inPubMed Google
Scholar * Damiana Francesca View author publications You can also search for this author inPubMed Google Scholar * Giulia Lorenzini View author publications You can also search for this
author inPubMed Google Scholar * Leonardo Barellini View author publications You can also search for this author inPubMed Google Scholar * Alfredo Falcone View author publications You can
also search for this author inPubMed Google Scholar * Alessandro Cosimi View author publications You can also search for this author inPubMed Google Scholar * Guido Bocci View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS G.A. and G.B. designed the research study. L.C., G.A., A.F., L.C.M., G.A., M.T.B., E.D.M., S.D., S.G.,
L.T., B.S., L.L., I.M., I.P., M.D.L., M.P., V.C., A.F., S.C., F.O., D.F., G.L., L.B., A.F., and A.C. performed the clinical research and collected the samples. P.O., M.B., E.F., P.G., and
G.F. conducted the laboratory experiments. M.S., P.O., and C.F. analyzed the data. L.C., G.A., C.F., and G.B. wrote the manuscript. All authors read and approve the final manuscript. L.C.,
G.A., and P.O. equally contributed to the present work. CORRESPONDING AUTHOR Correspondence to Guido Bocci. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION \LEMENTARY FILES REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use,
sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative
Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated
otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds
the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Coltelli, L., Allegrini, G., Orlandi, P. _et al._ A pharmacogenetic interaction analysis of bevacizumab with paclitaxel in advanced breast
cancer patients. _npj Breast Cancer_ 8, 33 (2022). https://doi.org/10.1038/s41523-022-00400-6 Download citation * Received: 17 June 2021 * Accepted: 07 February 2022 * Published: 21 March
2022 * DOI: https://doi.org/10.1038/s41523-022-00400-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative