Net-net auto machine learning (automl) prediction of complex ecosystems
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Biological Ecosystem Networks (BENs) are webs of biological species (nodes) establishing trophic relationships (links). Experimental confirmation of all possible links is difficult
and generates a huge volume of information. Consequently, computational prediction becomes an important goal. Artificial Neural Networks (ANNs) are Machine Learning (ML) algorithms that may
be used to predict BENs, using as input Shannon entropy information measures (Shk) of known ecosystems to train them. However, it is difficult to select _a priori_ which ANN topology will
have a higher accuracy. Interestingly, Auto Machine Learning (AutoML) methods focus on the automatic selection of the more efficient ML algorithms for specific problems. In this work, a
preliminary study of a new approach to AutoML selection of ANNs is proposed for the prediction of BENs. We call it the Net-Net AutoML approach, because it uses for the first time Shk values
of both networks involving BENs (networks to be predicted) and ANN topologies (networks to be tested). Twelve types of classifiers have been tested for the Net-Net model including linear,
Bayesian, trees-based methods, multilayer perceptrons and deep neuronal networks. The best Net-Net AutoML model for 338,050 outputs of 10 ANN topologies for links of 69 BENs was obtained
with a deep fully connected neuronal network, characterized by a test accuracy of 0.866 and a test AUROC of 0.935. This work paves the way for the application of Net-Net AutoML to other
systems or ML algorithms. SIMILAR CONTENT BEING VIEWED BY OTHERS MULTIPLE THRESHOLDS AND TRAJECTORIES OF MICROBIAL BIODIVERSITY PREDICTED ACROSS BROWNING GRADIENTS BY NEURAL NETWORKS AND
DECISION TREE LEARNING Article Open access 16 August 2021 A NOVEL HYBRID MODEL FOR SPECIES DISTRIBUTION PREDICTION USING NEURAL NETWORKS AND GREY WOLF OPTIMIZER ALGORITHM Article Open access
20 May 2024 A STUDY OF COMBINATION OF AUTOENCODERS AND BOOSTED BIG-BANG CRUNCH THEORY ARCHITECTURES FOR LAND-USE CLASSIFICATION USING REMOTELY SENSED IMAGERY Article Open access 02 May 2025
INTRODUCTION Many important molecular, living, economical, and other complex systems may be described as complex networks of _i_ parts or nodes interconnected by links, edges, bonds, ties,
or relationships1,2,3,4,5,6,7. The volume of information about all these collections of nodes and links is so large that it is impossible for a single person to remember and rationalize all
possible connections in known networks. Consequently, it is even more difficult to assign/predict correct connections in new cases. This problem can be solved using Machine Learning (ML)
models. In this area, ML models used as input variables are able to quantify structural information of the system. The process has been applied to multiple levels, ranging from the
prediction of drug-target networks in molecules to the construction of complex biological networks8,9,10,11. Specifically, a molecular or living complex system can be explained using
numerical parameters that quantify information about the structure of the system. In information theory, Shannon entropy quantifies the information contained in a message, usually in bits.
The concept was introduced by Claude E. Shannon in his 1948 paper “A Mathematical Theory of Communication”12. With the pass of time, the Shannon entropy information measures (Shk) of
different types and other related information measures have become commonly used indices in quantifying information of the system under study in ML
modelling13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29. In any case, developing ML models using as input Shk values involves, as in other ML problems, the application of data
pre-processing variable selection and other techniques. Next, it is necessary to _a priori_ select one or more ML algorithms and train/validate them to seek the final ML model. Consequently,
non-experts in ML may encounter difficulties to accomplish this goal. Specifically, in the case of complex molecular and living systems, a non-expert may find it difficult to decide a
priori which ML algorithms should be selected to develop the model. In this context, Automated Machine Learning (AutoML) may have an important role in automatically selecting ML algorithms
during the development of practical ML applications by non-experts30,31. This work proposes for the first time the use of Shk values to quantify both the structure of the complex biological
system to be predicted and the structure of the ML algorithm to be selected for this task. To this end, a preliminary proof-of-concept experiment is carried out, focusing on a specific class
of complex biological systems, and a specific type of ML algorithms. Biological Ecosystem Networks (BENs) have been selected to play the role of a complex biological system. In addition,
Artificial Neural Networks (ANN) have been selected to represent the ML algorithms. The current study uses the entropy values Shk(Ai) and Shk(Bj) as inputs for different pairs of species in
the BENs, of the systems under study. The Shk(ANNj) values calculated are also used as inputs for different ANN topologies. In fact, Ecosystems represent one of the most important examples
of complex systems. They are a clear example of network-like structures with known procedures to calculate the Shk values32,33,34. In this sense, our group reported different ML models that
evaluate the structure of parasite-host webs to predict the interactions between species in different networks35,36,37. In one of our previous works, special emphasis has been placed on the
use of Shk information measures to codify structural information in this type of ML studies38. On the other hand, ANNs have been selected because of their more apparent network-like
structure, and because they are a useful tool to solve this kind of ML problems. In fact, ANNs are powerful bio-inspired algorithms able to learn/infer large datasets39,40,41,42. ANNs are
also able to learn topology patterns in large datasets of bio-systems and other complex networks43. This work proposes the calculation of Shk information indices in both sets of networks:
BENs and ANNs. That is why, it has been called a Net-Net AutoML approach. Last, an AutoML linear model is sought using these indices as input. This Net-Net AutoML model could be employed to
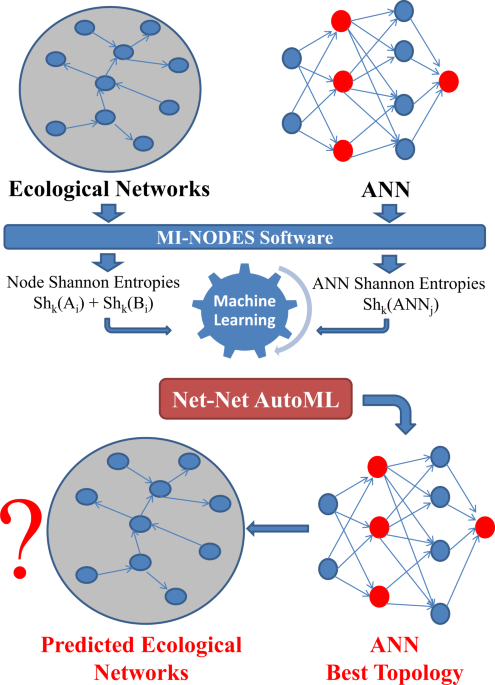
screen different ANN topologies in order to pre-select the one expected to correctly predict BEN structures before training it. RESULTS This work introduces for the first time a new type of
algorithm to find the best ANNs that predict BENs. The main steps of the methodology are described in Fig. 1. This is the first report of a Net-Net AutoML model for ANN screening, with the
subsequent saving of time and computational resources in the prediction of Complex Networks. The BEN node pairs and the ANN classifiers that were trained for the prediction of BEN node
connectivity were turned into Shk descriptors that encoded information for the BEN nodes and the entire ANN topology. Shk were calculated for each node with the MI-NODES software44. In the
case of ANN classifiers, the average of all the values of Shk for all the neurons in the ANN was used as input. For the MI-NODES descriptors, the Markov chains theory was applied and,
therefore, they were calculated for each _k_ values ranging between 0 and 5 (_k_ = node distance of interaction) as Shk45. These descriptors were linearly combined to find a model (AutoML)
that was able to predict how a specific ANN topology would evaluate BEN node connectivity. Thus, AutoML could be used to screen which is the best ANN classifier topology for BEN node
connectivity prediction. The AutoML methodology used for the prediction of BEN connectivity includes the following steps with their respective results. First, Shk values were calculated for
a large number of nodes in 69 BENs using the MI-NODES software. We created a dataset of biological systems (bsi dataset) using 33,805 pairs of nodes selected randomly from the 69 BENs. If we
consider the adjacency matrix (A) as the mathematical representation of all pairs of A_i_ vs. B_j_ nodes in the BEN, the output variable of this dataset are the elements Aij of this matrix.
These values quantify the structure (connectivity) of the BEN with values Aij = 1 for the pairs of nodes that are connected (interacting biological species) and Aij = 0 otherwise
(non-interacting biological species). Next, the bsi dataset was expanded with node differences as input variables ∆Shkij = Shki − Shkj for each pair, where Shki is the Shk for the first node
and Shkj the Shk for the second node (k = 0–5). As a result, there are bsiNvar = 6·3 = 18 input features for the Aij output for each pair of BEN nodes. The variables Sh0 quantify
information for an isolated node, Sh1 refers to the nodes with direct link, Sh2 to nodes that have other nodes between them, and so on. Figure 2 illustrates the distribution of three Shk
parameters (k = 2, 3, 5) for both BENs and ANN classifiers to predict them. This dataset was used to train 10 different ANNs. Next, the ANN screening model testing dataset (mt dataset) was
made up. The output variable of the mt dataset represents the values of correct or incurred prediction of BEN connectivity Aij by a specific ANN classifier topology, P(ANNAij) = 1 when the
ANN topology correctly predicts the observed BEN nodes connectivity Aij (Aij = 1 or 0 in the original bsi dataset). On the contrary, P(ANNAij) = 0 when a specific ANN topology fails to
correctly classify the observed Aij of 1 or 0 from the original bsi dataset. The mt dataset contains the predictions of 338,050 node pairs from 69 BENs using 10 different trained ANN
classifiers (different topologies). The input variables of the mt dataset are the original variables for each pair of nodes and the values of information indices ANNShk (average value of Sh
of all ANN neurons): mtNvar = 6*(Shki + Shkj + ∆Shkij + ANNShk) = 24. The last step consists of the dataset analysis to find the best linear AutoML model for ANN classifier screening (see
previous Fig. 1). DISCUSSION There are at least two major problems if ANNs are used to predict node connectivity in complex networks. First, the information in complex systems should be
turned into numerical input parameters to the future ANN classifiers for node connectivity. Secondly, many ANN classifiers with different topologies should be trained in order to find the
best ANN topology that can learn the complex system structure patterns. The first problem can be solved by quantifying the structural information of the complex system (Brain, Ecological,
Social, etc.) with Shk information measures46. The classical solution for the second problem is the training of different ANNs to find the best topology. This step involves the use of High
Performance Computing (HPC) services if the aim is to test a high number of ANNs for many complex systems. The current study proposes a new methodology to evaluate how a new ANN classifier
could predict the BEN node connectivity, without the need for ANN training. Thus, two types of information descriptors were used: node descriptors for BEN complex networks and the average of
ANN neuron descriptors. If ANNs are networks with nodes (neurons) and links (weights), the same mathematical processing as in the case of the BEN complex network could be applied.
Therefore, it is possible to quantify topological (connectivity) information of both the BEN complex networks under study and a set of ANNs trained using Shk descriptors. Thus, each node of
the complex networks encoded information into Shk descriptors and each ANN classifier was characterized as an entire network by the average of Shk. The new AutoML methodology proposed a
screening of ANN classifier topologies for BEN node connectivity prediction. The current work applied the AutoML methodology to the Ecological systems. Consequently, the AutoML output
P(ANNAij) predicted the propensity of a specific ANN topology to predict the biological interaction Aij between the species A_i_ and B_j_ from the ecological web (Aij = 1 or not Aij = 0).
The best linear AutoML with maximum values of Ac, Sp, and Sn (training and external validation series) is described by Eq. 1. The best linear AutoML was made up of only 5 features: two Shk
(_k_ = 0, 3) for each node A_i_ and B_j_, and a Shk (_k_ = 3) for the ANN classifier. $$\begin{array}{rcl}{\rm{S}}({\rm{L}}={1,\mathrm{ANN}}_{{\rm{j}}})\, & = &
-{\rm{42}}\mathrm{.38}\cdot S{h}_{0}({A}_{i})+15.69\cdot S{h}_{3}({A}_{i})\\ & & -{\rm{44.95}}\cdot S{h}_{0}({B}_{j})+13.79\cdot S{h}_{3}({B}_{j})\\ & & -{\rm{0.014}}\cdot
S{h}_{3}(AN{N}_{j})-0.8263\\ & & {\rm{n}}=235\,\,540\,{\chi }^{2}\,=\mathrm{56326}{\rm{.2}}\,p < 0{\rm{.001}}\end{array}$$ (1) In each BEN, the connections (Aij = 1) indicated the
existence of a biological interaction between the organisms of _i_ biological species with the organisms of _j_ species. Table 1 shows the ANN topologies trained to predict BEN
connectivity. Thus, the Net-Net AutoML model was able to predict whether a new ANN topology could correctly predict the connectivity between a pair of BEN nodes, prior to training. We
introduced variability in the ANN topologies using ANNs without hidden layers (no. 8, 9, 10), with only one hidden layer (no. 1, 2, 7, 5) and with two hidden layers (no. 3, 4, 6). Future
work should include different ANN topologies such as skip layer connections, drop out neurons, deep ANNs. These will enable a wider search in the space of possible networks. The LDA model
showed significant goodness-of-fit, also illustrated by Accuracy (Ac), Sensitivity (Sn), and Specificity (Sp) classification values, both in training and external validation series (see
Table 2). The proof-of-concept AutoML model fit very well 338,050 outcomes predicted with 10 (previously trained) ANNs. These results were obtained after training the 10 ANNs to learn to
discriminate between biological interactions (predation, parasitism, mutualism, _etc_.) which were connected (Aij = 1) or not (Aij = 0) in BENs of many ecosystems. The mission of the AutoML
did not consist of the prediction of BEN connectivity, and, therefore, Sn referred to the number of times that the AutoML was able to evaluate whether a given ANN topology could correctly
predict BEN nodes connectivity. The same analogy applied to Sp and Ac. Using Net-Net AutoML methodology, one could decide which ANN will receive more computing resources for training and
which one can be used to predict different links (Aij = 1 or 0). The parameter Sh3i quantified the information related to the position of _i_ organism and their neighbours (_k_ = 3) placing
a topological distance d ≤ 3 in the BEN. ANNSh3 is also similar but quantifies information for the neurons in a specific ANN topology and not for the organisms in the biological network.
Figure 2 illustrates the distribution of the Shk values for all the BENs and ANN topologies studied herein. The LDA model is a base line classifier that was compared to 11 complex
classifiers obtained with 9 ML methods, such as Bayesian Nets, Naïve Bayes Nets, Logistic Regression, Decision Table, Multilayer Perceptron (MLP), Random Forest, Bagging, AdaBoost, and Deep
Fully Connected (FC) Networks. All 18 descriptors were used as inputs. The test accuracy (ACC) and AUROC values are presented in Table 3. It should be observed that the Bayesian methods,
Decision Table and Logistic Regression provided accuracies lower than the LDA model. With the MLP, by introducing hidden layers in Artificial Neural Networks, the accuracies and AUROC were
improved, with values over 0.8, better than the LDA classifier. More complex models such as ensemble classifiers based on MLP with only one hidden layer (Bagging MLP and AdaBoostM1 MLP)
could produce slightly better results. By introducing more hidden layers with MLP 2 H and Deep FC Nets, the test accuracy was increased up to 0.866 (test AUROC = 0.935). Random Forest was
not the best model, but it was able to provide a test accuracy of 0.832 (AUROC = 0.914). The ensemble classifier based on simple REP trees, such as Bagging REP, had a performance similar to
the MLP 1 H (only one hidden layer). Therefore, Deep Nets provided the best results, starting with MLP 2H with only 18 neurons (=number of input features) in the first hidden layer, and 9
neurons in the second hidden layer (ACC = 0.827, AUROC = 0.902), leading to the more complex Deep FC Nets with 200, 400 and 200 neurons in the hidden layers 1, 2 and 3 (ACC = 0.866, AUROC =
0.935). An accuracy increase of 4% was obtained with more complex topology of the neural network from 18–9 to 200–400–200 neurons. The DL model was obtained using 10 different network
topologies, from one to three hidden layers, with different optimization algorithms, dropout rates, and other hyperparameters. The best DL model had the hidden layer topology n-n*2-n, with
activation functions = tanh, n = 200, dropout rate = 0.5, optimizer algorithm = Adam, initialization of weights = glorot_normal, batch size = 4096, epochs = 500, training AUROC = 0.963,
training ACC = 0.897, test AUROC = 0.935 and test ACC = 0.866. The current method used different applications such as MI-NODES for descriptors, STATISTICA, Weka, and Python/Keras for ML
classifiers. If the user does not test deep learning classifiers for the final Net-Net model, there is no need for programming. In Weka it is possible to test a deeper MLP. Therefore,
scientists without advanced knowledge of programming are able to implement this methodology for specific BENs. An optimal implementation of the method should be performed using a unique
python code for all the Net-Net methodology steps. This is the next step for the future version of this method. CONCLUSIONS The current study confirms that Markov chains are useful to
calculate Shk information indices in order to quantify the connectivity patterns of both BENs and ANNs. The new Net-Net AutoML methodology demonstrated how to develop a linear AutoML model,
able to select which ANN topology would correctly predict the connectivity of BEN nodes before training it. The best AutoML model demonstrated an accuracy over 86% in test subsets. In
conclusion, Net-Net AutoMLs with Shk information indices could be used to screen ANN topologies that can predict the links in biological networks. This may lead to an optimization of
computing resources with the prioritization of the training of the best ANN topologies. METHODS BIOLOGICAL ECOSYSTEM NETWORKS DATASET A number of 69 Ecosystems or Food Webs were used. The
network files in.net format were assembled by our group in a previous work47. The datasets were downloaded from the Interaction Web Database (IWDB):
http://www.nceas.ucsb.edu/interactionweb/index.html. COMPUTATIONAL MODEL MARKOV-SHANNON ENTROPY CENTRALITIES FROM MI-NODES TOOL In the present work, the classical Markov matrix (1Π) was
constructed for each network (BEN complex networks and ANNs). In the case of BENs, the adjacency/connectivity matrix were downloaded from public resources as A (_n_ by _n_ matrix, where _n_
is the number of nodes/vertices). Next, the Markov matrix Π was calculated. It contains the vertices probability (_p__ij_) based on A. The probability matrix was raised to the power _k_,
resulting (1Π)k, and it was multiplied by the vector of the initial probabilities (0_p__j_). The resulting vectors kP contained the absolute probabilities to reach the nodes moving
throughout a walk of length _k_ from node _j_ (_k__p__j_) for each _k_ (Eqs 2 and 3). The entropy of graph Shk(G) could be calculated based on the entropy of each node Shkj:
$$\,{}^{k}P=\,{}^{0}P\times {({}^{1}{\rm{\Pi }})}^{k}=[\,{}^{k}p_{1},\,{}^{k}p_{2},\,\ldots ,\,{}^{k}p_{j}]$$ (2) $$S{h}_{k}(G)=\sum _{j\in G}S{h}_{kj}=-\,\sum _{j\in
G}\,{}^{k}p_{j}\,\mathrm{log}{}^{k}p_{j}$$ (3) NET-NET AUTOML MODELS Due to the dimension of the dataset and the complexity of the models, for the current calculations two systems were used:
* BioCAI cluster of RNASA-IMEDIR group (UDC) with 200 CPU cores; * A desktop computer with processor i7 (3.60 GHz × 4 physical cores), 16 G RAM, and a GPU NVIDIA Titan Xp (Pascal
architecture, dedicated memory of 12 G G5X, 3840 CUDA cores, boost clock 1,582 MHz). The GPU was particularly useful for the Deep Learning calculations with Keras. All the calculations could
be carried out with a desktop computer over a larger period a time, especially due to the Deep Learning calculations. Once the Shk values of both the ANNs and BENs have been obtained, a
Linear Discriminant Analysis (LDA) implemented in the STATISTICA software48 can be run. Let P(ANNAij) be the output of a screening model used to predict the ability of a given ANN topology
to correctly classify the BEN connectivity Aij between two nodes _i_ and _j_ (Aij = 1 or 0). Eq. 4 describes the general formula for the LDA model using the following coefficients: aki as
coefficients of the Sh for the _i_ node (Shki), bkj as coefficients of the Sh for the _j_ node (Shkj), ckij as coefficients of the differences between the Sh of the nodes _i_ and _j_ (∆Shkij
= Shki − Shkj), ANNdk as coefficient of the average Sh for a specific ANN topology, and e0 as the free term coefficient. The _k_ index indicates that this Shk value codifies information for
all nodes placed at least at topological distance _k_ from the reference node. Different statistical parameters can be used to evaluate the statistical significance and validate the
goodness-of-fit of LDA equation: n = number of cases, χ2 = Chi-square, p = error level, as well as the Accuracy (Ac), Specificity (Sp), and Sensitivity (Sn) of both training and external
validation series49. $$S(L=1,ANN)=\sum _{k=0}^{5}{a}_{ki}\cdot S{h}_{ki}+\sum _{k=0}^{5}{b}_{kj}\cdot S{h}_{kj}+\sum _{k=0}^{5}{c}_{kij}\cdot {\rm{\Delta }}S{h}_{kij}+\sum
_{k=0}^{5}{}^{ANN}d_{k}\cdot {}^{ANN}S{h}_{k}+{e}_{0}$$ (4) Several complex classifiers were tested (see Table 3): * Bayesian Nets = weka.classifiers.bayes.BayesNet -D -Q
weka.classifiers.bayes.net.search.local.K2–P 1 -S BAYES -E weka.classifiers.bayes.net.estimate.SimpleEstimator –A 0. * Naive Bayes Nets = weka.classifiers.bayes.NaiveBayes * Logistic
Regression = weka.classifiers.functions.Logistic -R 1.0E-8 -M -1 -num-decimal-places 4 * Decision Table = weka.classifiers.rules.DecisionTable -X 1 -S “weka.attributeSelection.BestFirst -D 1
-N 5” * MLP 1H (Multilayer Perceptron 1 hidden layer) = weka.classifiers.functions.MultilayerPerceptron -L 0.3 -M 0.2 -N 1000 -V 0 -S 0 -E 20 -H a * MLP 2H (Multilayer Perceptron 2 hidden
layers) = weka.classifiers.functions.MultilayerPerceptron -L 0.3 -M 0.2 -N 5000 -V 0 -S 0 -E 20 -H “18, 9” -batch-size 500 * Random Forest = weka.classifiers.trees.RandomForest -P 100 -I 500
-num-slots 1 -K 0 -M 1.0 -V 0.001 -S 1 * Bagging REP = weka.classifiers.meta.Bagging -P 100 -S 1 -num-slots 1 -I 10 -W weka.classifiers.trees.REPTree –M 2 -V 0.001 -N 3 -S 1 -L -1 -I 0.0 *
Bagging MLP = weka.classifiers.meta.Bagging -P 100 -S 1 -num-slots 1 -I 10 -W weka.classifiers.functions.MultilayerPerceptron -batch-size 4000 –L 0.3 -M 0.2 -N 500 -V 0 -S 0 -E 20 -H a *
AdaBoostM1 MLP = weka.classifiers.meta.AdaBoostM1 -P 100 -S 1 -I 10 -W weka.classifiers.functions.MultilayerPerceptron -batch-size 4000 –L 0.3 -M 0.2 -N 500 -V 0 -S 0 -E 20 -H a * Deep FC
Nets (Deep Learning Fully Connected Networks) = n-n*2-n′ hidden layer topology (n = 200). Deep Learning FC Nets were programmed in Python with Keras, and the other classifiers were obtained
with the Weka tool. For the DL models, different hyperparameter values were tested: * n = Number of neurons in a hidden layer: 10, 18, 50, 100, 200, 500. * Network topologies: ‘n’,‘n-n’,
‘n-n-n’, ‘n-n*2’, ‘n-n*2-n’, ‘n*2’, ‘n*2-n’, ‘n-n:2’, ‘n:2’, ‘n-n:2-n:4’ (this notation does not include the input layer with 18 neurons = no. of features and the output layer with a neuron
for the class). * Neuron dropout rate: 0.0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9. * Optimizers: ‘RMSprop’, ‘Adagrad’, ‘Adadelta’, ‘Adam’, ‘Adamax’, ‘Nadam’. * Weight initialization
for hidden layer neurons: ‘uniform’, ‘lecun_uniform’, ‘normal’, ‘glorot_normal’, ‘glorot_uniform’, ‘he_normal’, ‘he_uniform’. * Batch size for training = 1024, 2048, 4096. * Training epochs
= 20, 50, 100, 200, 300, 400, 500. * Training cross validation: 3-folds (default value in Keras). The Net-Net AutoML algorithm shown in Fig. 1 could be described as follows: * (1) For each
BEN: * (1.1) Get the connectivity matrix. * (1.2) Add weights for the BEN connections (if present). * (1.3) For each node _A_: * (1.3.1) Calculate node Shannon Entropies with MI-NODES:
Shk(Ai). * (1.3.2) Create pairs of entropies for all the other nodes _B_: Shk(Ai) − Shk(Bj). * (2) Find different ANN classifiers to predict BEN node Aj − Bj connectivity: * (2.1) For each
ANNj classifier: * (2.1.1) Calculate network Shannon Entropy: Shk(ANNj). * (3) Merge BEN node descriptors with ANN descriptors into Net-Net dataset: Shk(Ai), Shk(Bj), Shk(ANNj). * (4) Split
Net-Net dataset into training and test subsets. * (5) Find the best Net-Net classifier to evaluate whether a specific ANN can predict the BEN connectivity: * (5.1) For each ML method
(Bayesian, Trees, Artificial Neural Networks, etc.) * (5.1.1) For each set of model parameters (ex: topology, activation function, etc.) * (5.1.1.1) Use a Net-Net subset to train the
classifier * (5.1.1.2) Evaluate the model with test subset calculating accuracy (ACC) and AUROC. * (6) Choose the best Net-Net classifier with the best ACC and AUROC. Steps (5) and (6) used
Weka and Python/Keras scripts. In the future version of the method, different classifiers will be tested for the BEN connectivity prediction (not only ANNs). This involves the adaptation of
MI-NODES application. The main advantage of the Net-Net methodology is that it can build a Net-Net classifier able to screen ANN classifiers which predict BEN node connectivities. DATA
AVAILABILITY All data generated or analyzed during this study were included in this article (along with its Supplementary Information files) and they are publicly available at Figshare
repository with https://doi.org/10.6084/m9.figshare.6238424. REFERENCES * Sandhu, K. S. _et al_. Large-scale functional organization of long-range chromatin interaction networks. _Cell Rep_
2, 1207–1219, https://doi.org/10.1016/j.celrep.2012.09.022 (2012). Article PubMed PubMed Central CAS Google Scholar * Gaspar, M. E. & Csermely, P. Rigidity and flexibility of
biological networks. _Brief Funct Genomics_ 11, 443–456, https://doi.org/10.1093/bfgp/els023 (2012). Article PubMed Google Scholar * Csermely, P., Korcsmaros, T., Kiss, H. J., London, G.
& Nussinov, R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. _Pharmacol. Ther._ 138, 333–408,
https://doi.org/10.1016/j.pharmthera.2013.01.016 (2013). Article PubMed PubMed Central CAS Google Scholar * Vidal, M., Cusick, M. E. & Barabasi, A. L. Interactome networks and human
disease. _Cell_ 144, 986–998, https://doi.org/10.1016/j.cell.2011.02.016 (2011). Article PubMed PubMed Central CAS Google Scholar * Barabasi, A. L., Gulbahce, N. & Loscalzo, J.
Network medicine: a network-based approach to human disease. _Nat Rev Genet_ 12, 56–68, https://doi.org/10.1038/nrg2918 (2011). Article PubMed PubMed Central CAS Google Scholar *
Barabasi, A. L. & Oltvai, Z. N. Network biology: understanding the cell’s functional organization. _Nat Rev Genet_ 5, 101–113, https://doi.org/10.1038/nrg1272 (2004). Article PubMed
CAS Google Scholar * Strogatz, S. H. Exploring complex networks. _Nature_ 410, 268–276, https://doi.org/10.1038/35065725 (2001). Article ADS PubMed MATH CAS Google Scholar *
Riera-Fernandez, P. _et al_. From QSAR models of Drugs to Complex Networks: State-of-Art Review and Introduction of New Markov-Spectral Moments Indices. _Curr Top Med Chem_ 12, 927–960,
https://doi.org/10.2174/156802612800166819 (2012). Article PubMed CAS Google Scholar * Gonzalez-Diaz, H. QSAR and Complex Networks in Pharmaceutical Design, Microbiology, Parasitology,
Toxicology, Cancer and Neurosciences. _Current Pharmaceutical Design_ 16, 2598–U2524, https://doi.org/10.2174/138161210792389261 (2010). Article PubMed CAS Google Scholar *
González-Díaz, H., Prado-Prado, F., Pérez-Montoto, L. G., Duardo-Sánchez, A. & López-Díaz, A. QSAR Models for Proteins of Parasitic Organisms, Plants and Human Guests: Theory,
Applications, Legal Protection, Taxes, and Regulatory Issues. _Curr Proteomics_ 6, 214–227, https://doi.org/10.2174/157016409789973789 (2009). Article Google Scholar * Prado-Prado, F. J.,
Ubeira, F. M., Borges, F. & Gonzalez-Diaz, H. Unified QSAR & Network-Based Computational Chemistry Approach to Antimicrobials. II. Multiple Distance and Triadic Census Analysis of
Antiparasitic Drugs Complex Networks. _J. Comput. Chem._ 31, 164–173, https://doi.org/10.1002/jcc.21292 (2010). Article PubMed CAS Google Scholar * Shannon, C. E. A Mathematical Theory
of Communication. _The Bell System Technical Journal_ 27, 379–423, https://doi.org/10.1002/j.1538-7305.1948.tb01338.x (1948). Article MathSciNet MATH Google Scholar * Dehmer, M. &
Emmert-Streib, F. Analysis of Complex Networks. _From Biology to Linguistics_. (WILEY-VCH Verlag GmbH & Co. KGaA, 2009). * Dehmer, M., Grabner, M. & Varmuza, K. Information indices
with high discriminative power for graphs. _PLoS ONE_ 7, e31214, https://doi.org/10.1371/journal.pone.0031214 (2012). Article ADS PubMed PubMed Central CAS Google Scholar * Dehmer, M.,
Varmuza, K., Borgert, S. & Emmert-Streib, F. On entropy-based molecular descriptors: statistical analysis of real and synthetic chemical structures. _Journal of chemical information and
modeling_ 49, 1655–1663 (2009). Article PubMed CAS Google Scholar * Estrada, E. & Avnir, D. Continuous symmetry numbers and entropy. _J Am Chem Soc_ 125, 4368–4375,
https://doi.org/10.1021/ja020619w (2003). Article PubMed CAS Google Scholar * Graham, D. J., Grzetic, S., May, D. & Zumpf, J. Information properties of naturally-occurring proteins:
Fourier analysis and complexity phase plots. _The protein journal_ 31, 550–563, https://doi.org/10.1007/s10930-012-9432-7 (2012). Article PubMed CAS Google Scholar * Graham, D. J. &
Greminger, J. L. On the information expressed in enzyme structure: more lessons from ribonuclease A. _Mol. Divers._ 15, 769–779, https://doi.org/10.1007/s11030-011-9307-4 (2011). Article
PubMed CAS Google Scholar * Graham, D. J. & Greminger, J. L. On the information expressed in enzyme primary structure: lessons from Ribonuclease A. _Mol. Divers._ 14, 673–686,
https://doi.org/10.1007/s11030-009-9211-3 (2010). Article PubMed CAS Google Scholar * Graham, D. J. & Kim, M. Information and classical thermodynamic transformations. _The journal of
physical chemistry_ 112, 10585–10593, https://doi.org/10.1021/jp7119526 (2008). Article PubMed CAS Google Scholar * Graham, D. J., Malarkey, C. & Sevchuk, W. Experimental
investigation of information processing under irreversible Brownian conditions: work/time analysis of paper chromatograms. _The journal of physical chemistry_ 112, 10594–10602,
https://doi.org/10.1021/jp711953r (2008). Article PubMed CAS Google Scholar * Graham, D. J. Information Content in Organic Molecules: Brownian Processing at Low Levels. _Journal of
chemical information and modeling_ 47, 376–389 (2007). Article PubMed CAS Google Scholar * Graham, D. J. Information content in organic molecules: aggregation states and solvent effects.
_Journal of chemical information and modeling_ 45, 1223–1236, https://doi.org/10.1021/ci050101m (2005). Article PubMed CAS Google Scholar * Graham, D. J. & Schulmerich, M. V.
Information Content in Organic Molecules: Reaction Pathway Analysis via Brownian Processing. _J Chem Inf Comput Sci_ 44 (2004). * Graham, D. J., Malarkey, C. & Schulmerich, M. V.
Information Content in Organic Molecules: Quantification and Statistical Structure via Brownian Processing. _J. Chem. Inf. Comput. Sci_. 44 (2004). * Graham, D. J. Information and organic
molecules: structure considerations via integer statistics. _J. Chem. Inf. Comput. Sci._ 42, 215–221 (2002). Article PubMed CAS Google Scholar * Graham, D. J. & Schacht, D. V. Base
information content in organic formulas. _J. Chem. Inf. Comput. Sci._ 40, 942–946 (2000). Article PubMed CAS Google Scholar * Barigye, S. J. _et al_. Shannon’s, Mutual, Conditional and
Joint Entropy Information Indices. Generalization of Global Indices Defined from Local Vertex Invariants. _Curr Comput Aided Drug Des_ (2013). * Aguiar-Pulido, V. _et al_. Naïve Bayes QSDR
classification based on spiral-graph Shannon entropies for protein biomarkers in human colon cancer. _Mol Biosyst_, https://doi.org/10.1039/c2mb25039j (2012). * Kotthoff, L., Thornton, C.,
Hoos, H. H., Hutter, F. & Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. _Journal of Machine Learning Research_ 18, 1–5 (2017). *
Feurer, M., Klein, A., Eggensperger, K., Springenberg, J. & Blum, M. Efficient and Robust Automated Machine Learning. _Advances in Neural Information Processing Systems_ 28, 2962–2970
(2015). Google Scholar * Borenstein, E. & Feldman, M. W. Topological signatures of species interactions in metabolic networks. _J Comput Biol_ 16, 191–200,
https://doi.org/10.1089/cmb.2008.06TT (2009). Article PubMed PubMed Central CAS Google Scholar * Ulanowicz, R. E. Quantitative methods for ecological network analysis. _Comput Biol
Chem_ 28, 321–339, https://doi.org/10.1016/j.compbiolchem.2004.09.001 (2004). Article PubMed MATH CAS Google Scholar * Olff, H. _et al_. Parallel ecological networks in ecosystems.
_Philos Trans R Soc Lond B Biol Sci_ 364, 1755–1779, https://doi.org/10.1098/rstb.2008.0222 (2009). Article PubMed PubMed Central Google Scholar * Gonzalez-Diaz, H., Riera-Fernandez, P.,
Pazos, A. & Munteanu, C. R. The Rucker-Markov invariants of complex Bio-Systems: applications in Parasitology and Neuroinformatics. _Biosystems_ 111, 199–207,
https://doi.org/10.1016/j.biosystems.2013.02.006 (2013). Article PubMed Google Scholar * Gonzalez-Diaz, H. & Riera-Fernandez, P. New Markov-Autocorrelation Indices for Re-evaluation
of Links in Chemical and Biological Complex Networks used in Metabolomics, Parasitology, Neurosciences, and Epidemiology. _J. Chem. Inf. Model._ 52, 3331–3340,
https://doi.org/10.1021/ci300321f (2012). Article PubMed CAS Google Scholar * Riera-Fernandez, I. _et al_. From QSAR models of Drugs to Complex Networks: State-of-Art Review and
Introduction of New Markov-Spectral Moments Indices. _Curr. Top. Med. Chem_. (2012). * Riera-Fernandez, P. _et al_. New Markov-Shannon Entropy models to assess connectivity quality in
complex networks: From molecular to cellular pathway, Parasite-Host, Neural, Industry, and Legal-Social networks. _Journal of Theoretical Biology_ 293, 174–188,
https://doi.org/10.1016/j.jtbi.2011.10.016 (2012). Article PubMed Google Scholar * Gonzalez-Diaz, H. _et al_. ANN-QSAR model for selection of anticancer leads from structurally
heterogeneous series of compounds. _European Journal of Medicinal Chemistry_ 42, 580–585, https://doi.org/10.1016/j.ejmech.2006.11.016 (2007). Article PubMed CAS Google Scholar *
Jalali-Heravi, M. & Fatemi, M. H. Prediction of thermal conductivity detection response factors using an artificial neural network. _J. Chromatogr. A_ 897, 227–235 (2000). Article
PubMed CAS Google Scholar * Prado-Prado, F. J., Garcia-Mera, X. & Gonzalez-Diaz, H. Multi-target spectral moment QSAR versus ANN for antiparasitic drugs against different parasite
species. _Bioorganic & Medicinal Chemistry_ 18, 2225–2231, https://doi.org/10.1016/j.bmc.2010.01.068 (2010). Article CAS Google Scholar * Tenorio-Borroto, E. _et al_. ANN multiplexing
model of drugs effect on macrophages; theoretical and flow cytometry study on the cytotoxicity of the anti-microbial drug G1 in spleen. _Bioorganic & Medicinal Chemistry_ 20, 6181–6194,
https://doi.org/10.1016/j.bmc.2012.07.020 (2012). Article CAS Google Scholar * Gonzalez-Diaz, H. _et al_. MIANN models in medicinal, physical and organic chemistry. _Curr Top Med Chem_
13, 619–641 (2013). Article PubMed CAS Google Scholar * Duardo-Sanchez, A. _et al_. Modeling complex metabolic reactions, ecological systems, and financial and legal networks with MIANN
models based on Markov-Wiener node descriptors. _Journal of chemical information and modeling_ 54, 16–29, https://doi.org/10.1021/ci400280n (2014). Article PubMed CAS Google Scholar *
Duardo-Sanchez, A., Gonzalez-Diaz, H. & Pazos, A. MI-NODES Multiscale Models of Metabolic Reactions, Brain Connectome, Ecological, Epidemic, World Trade, and Legal-Social Networks.
_Curr. Bioinf._ 10, 692–713, https://doi.org/10.2174/1574893610666151008013413 (2015). Article CAS Google Scholar * Shannon, C. E., Weaver, W., Blahut, R. E. & Hajek, B. _The
mathematical theory of communication_. Vol. 117 (University of Illinois press Urbana, 1949). * Riera-Fernández, P. _et al_. Definition of Markov-Harary Invariants and Review of Classic
Topological Indices and Databases in Biology, Parasitology, Technology, and Social-Legal Networks. _Current Bioinformatics_ 6, 94–121 (2011). Article Google Scholar * STATISTICA (data
analysis software system), version 6. 0, www.statsoft.com.Statsoft, Inc. v. 6.0 (2002). * Hill, T. & Lewicki, P. _STATISTICS Methods and Applications. A Comprehensive Reference for
Science, Industry and Data Mining_. Vol. 1 (StatSoft, 2006). Download references ACKNOWLEDGEMENTS The authors acknowledge Basque Government (Eusko Jaurlaritza) grant (IT1045-16) - 2016–2021
for consolidated research groups. This work was supported by the “Collaborative Project in Genomic Data Integration (CICLOGEN)” PI17/01826 funded by the Carlos III Health Institute, as part
of the Spanish National plan for Scientific and Technical Research and Innovation 2013–2016 and the European Regional Development Funds (FEDER). This project was also supported by the
General Directorate of Culture, Education and University Management of Xunta de Galicia ED431D 2017/16 and “Drug Discovery Galician Network” Ref. ED431G/01 and the “Galician Network for
Colorectal Cancer Research” (Ref. ED431D 2017/23), and finally by the Spanish Ministry of Economy and Competitiveness for its support through the funding of the unique installation BIOCAI
(UNLC08-1E-002, UNLC13-13-3503) and the European Regional Development Funds (FEDER) by the European Union. CR Munteanu acknowledges the support of NVIDIA Corporation with the donation of the
Titan Xp GPU used for this research. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Computation, Computer Science Faculty, University of A Coruna (UDC), 15071, A Coruña, Spain
Enrique Barreiro & Cristian R. Munteanu * Center for Computational Science (CCS), University of Miami (UM), Miami, 33136, FL, USA Enrique Barreiro & Maykel Cruz-Monteagudo * West
Coast University, Miami Campus, 33178, FL, USA Enrique Barreiro & Maykel Cruz-Monteagudo * Biomedical Research Institute of A Coruña (INIBIC), University Hospital Complex of A Coruña
(CHUAC), A Coruña, 15006, Spain Alejandro Pazos * Faculty of Science and Technology, University of the Basque Country (UPV/EHU), 48940, Biscay, Spain Humbert González-Díaz * IKERBASQUE,
Basque Foundation for Science, 48011, Bilbao, Biscay, Spain Humbert González-Díaz Authors * Enrique Barreiro View author publications You can also search for this author inPubMed Google
Scholar * Cristian R. Munteanu View author publications You can also search for this author inPubMed Google Scholar * Maykel Cruz-Monteagudo View author publications You can also search for
this author inPubMed Google Scholar * Alejandro Pazos View author publications You can also search for this author inPubMed Google Scholar * Humbert González-Díaz View author publications
You can also search for this author inPubMed Google Scholar CONTRIBUTIONS E.B. performed the data analysis and participated in the writing of the paper. C.R.M. coded MI-NODES scripts for the
calculation of Shk values and participated in the writing of the paper. M.C.M. supervised the work of E.B. on his internship and participated in the writing of the paper. A.P. contributed
to the discussion and participated in the writing of the paper. H.G.D. proposed the Net-Net AutoML algorithm idea, performed the data analysis, and participated in the writing of the paper.
CORRESPONDING AUTHOR Correspondence to Humbert González-Díaz. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S
NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION
RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Barreiro, E., Munteanu, C.R., Cruz-Monteagudo, M. _et al._ Net-Net Auto Machine Learning (AutoML) Prediction of Complex Ecosystems. _Sci Rep_ 8, 12340 (2018).
https://doi.org/10.1038/s41598-018-30637-w Download citation * Received: 09 May 2018 * Accepted: 24 July 2018 * Published: 17 August 2018 * DOI: https://doi.org/10.1038/s41598-018-30637-w
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative