Dnah11 variants and its association with congenital heart disease and heterotaxy syndrome
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Congenital heart diseases (CHDs) are the most common types of birth defects, affecting approximately 1% of live births and remaining the leading cause of mortality. CHD patients
often show a higher incidence of heterotaxy syndrome. However, the exact aetiology of CHD and heterotaxy syndrome remains unclear. In this study, targeted sequencing and Sanger sequencing
were performed to analyze the exonic regions of 37 primary ciliary dysfunction (PCD)- related candidate genes in 42 CHD patients with heterotaxy syndrome. Variants affecting protein-coding
regions were filtered according to databases of known variants and predicted in silico using functional prediction program. Thirty-four potential disease-causing heterozygous variants in 11
genes were identified in the 19 CHD patients with heterotaxy syndrome (45.2%, 19/42). The _DNAH11_ gene showed the highest mutation rate (16.7%; 14 of 84 alleles) among the CHD patients with
heterotaxy. Fisher’s exact test revealed a significant association of _DNAH11_ variants with CHD and heterotaxy (_P_ = 0.0001). In families, six different compound heterozygous variants of
_DNAH11_ were validated in family 1-5031 (p.W802X/p.M282I), family 2-5045 (p.T3460K/p.G4425S), family 3-5065 (p.G447R/p.L1157R), family 4-5130 (p.I2262T/p.D3800H), family 5-5707
(p.S1823fs/p.F2759L/p.R4395X) and family 6-5062 (p.D3610V/p.I243V). These findings suggest that the _DNAH11_ variants are significantly associated with CHD and heterotaxy syndrome and that
compound heterozygous _DNAH11_ variants may be the common genetic cause of the development of familial CHD and heterotaxy syndrome. SIMILAR CONTENT BEING VIEWED BY OTHERS THE PREVALENCE OF
LATERALITY DEFECTS IN PATIENTS WITH CONGENITAL HEART DISEASE Article 05 June 2025 THE C.1617DEL VARIANT OF _TMEM260_ IS IDENTIFIED AS THE MOST FREQUENT SINGLE GENE DETERMINANT FOR JAPANESE
PATIENTS WITH A SPECIFIC TYPE OF CONGENITAL HEART DISEASE Article Open access 26 February 2024 EXOME SEQUENCING IN INDIVIDUALS WITH CARDIOVASCULAR LATERALITY DEFECTS IDENTIFIES POTENTIAL
CANDIDATE GENES Article Open access 26 April 2022 INTRODUCTION Congenital heart diseases (CHDs) are the most common types of birth defects, affecting approximately 1% of live births and
remaining the leading cause of mortality1. Interestingly, CHD patients often show a higher incidence of heterotaxy syndrome. Studies have found that approximately 5–10% of CHD patients
present with heterotaxy syndrome2. Heterotaxy(HTX) is a rare birth defect involving left-right (LR) asymmetry with an incidence of 1 in 10,000 newborns, and approximately 90% of HTX patients
have complex CHDs3. CHD and heterotaxy syndrome have been shown to be associated with primary cilia dysfunction (PCD) or cilia dysfunction (CD). The mortality and respiratory complications
of CHD and heterotaxy in patients after cardiac surgery are significantly higher than those in the same type of patients without heterotaxy4. However, PCD is considered to be a monogenic
heterogeneous recessive disorder, while CHD and heterotaxy syndrome are multiple-gene complex inherited diseases5. The linkage between PCD and CHD /HTX needs to be investigated. Studies have
shown that mutations in genes causing PCD may be associated with the development of heterotaxy and/or CHD syndrome. Approximately 50% of PCD patients exhibit heterotaxy associated with
complex CHDs6. PCD-related genes in heterotaxy are thought to be responsible for the function of motile cilia in LR patterning, and CHD/heterotaxy patients also show increased airway CD
similar to that seen in PCD patients7. Researchers have shown that CHD and heterotaxy syndromes are multiple, complex, inherited diseases caused by numerous genes that are responsible for
inherited and sporadic cases8. Mutations in over 15 genes related to LR patterning have been observed in heterotaxy patients, but these findings account for fewer than 20% of heterotaxy
cases9. You Li _et al_. performed whole-exome sequencing in 218 CHD mouse models and identified 91 recessive CHD mutations in 61 genes, including 34 cilia-related genes, 16 genes involved in
cilia-transduced cell signalling, and 10 genes regulating vesicular trafficking (a pathway important for ciliogenesis and cell signaling), including a loss-of-function _DNAH11_ mutation
known to cause PCD10. _DNAH11_, located on chromosome 7p15.3, encodes a ciliary outer dynein arm protein (520 kDa) and is a member of the dynein heavy chain family, localizing exclusively to
the proximal region of respiratory cilia11. _DNAH11_ mutations have been found to result in abnormal ciliary ultrastructure and hyperkinetic ciliary beating12. Gene editing of _DNAH11_
mutations can restore normal cilia motility in primary ciliary dyskinesia13. An overwhelming majority of previous studies related to _DNAH11_ have focused on PCD and situs inversus totalis,
and few studies have concentrated on heterotaxy and CHD. To date, over 40 PCD-related genes have been found in heterotaxic patients with PCD. However, whether PCD-related gene mutations are
associated with CHD and heterotaxy syndrome remains unclear. In this study, we performed targeted sequencing and Sanger sequencing analysis of PCD-related genes in Chinese CHD patients with
heterotaxy syndrome to explore the genetic aetiology of CHD and heterotaxy syndrome. RESULTS ASSESSMENT OF THE CILIARY MOVEMENT STATUS IN CHD PATIENTS WITH HETEROTAXY SYNDROME A cohort of 42
CHD patients with heterotaxy syndrome were recruited from unrelated families, including 14 females (33.3%) and 28 males (66.7%) with ages ranging from 0.1 to 20.1 years (mean ± SD: 5.6 ±
4.7 years). To assess the ciliary movement status, we examined the ciliary beat pattern using a slow-motion playback of the video sequence to generate tracings of the ciliary beat and found
that 29 cases (69%, 29/42) showed CD (18 males and 11 females), and 13 cases (31%, 13/42) presented normal ciliary function (10 males and 3 females). The clinical features of the study
subjects are summarized in Table 1. TARGETED SEQUENCING ANALYSIS OF GENE VARIANTS IN CHD PATIENTS WITH HETEROTAXY SYNDROME To investigate the role of gene variants in CHD/heterotaxy
diseases, we carried out targeted sequencing on the exonic regions of the following 37 PCD-related candidate genes in 42 CHD patients with heterotaxy: _ABCC4_, _ARMC4_, _C21orf59_, _CCDC39_,
_CCDC40_, _CCDC65_, _CCDC114_, _CCDC151_, _CCNO_, _DNAAF1_, _DNAAF2_, _DNAFF3_, _DNAAF5_, _DNAH5_, _DNAH8_, _DNAH11_, _DNAI1_, _DNAI2_, _DNAL1_, _DRC1_, _DYX1C1_, _HEATR2_, _HYDIN_,
_LRRC6_, _NAT10_, _NME8_, _PTGES_, _PTGES2_, _PTGES3_, _PTGER4_, _PTGS1_, _PTGS2_, _RSPH1_, _RSPH4A_, _RSPH9_, _SPAG1_ and _ZMYND10_. For these genes, 758 regions of gene coding exons and
their intron/exon junctions were sequenced. The coverage of the sequencing results was 90%-99%, with an average coverage of approximately 97%, and the average depth of sequencing was 100X.
By filtering from 1000 Genomes Project and ExAC databases and using SIFT, PolyPhen2 and MutationTaster prediction programs, we focused on novel or rare coding variants (MAF < 0.01%)
present in CHD patients with heterotaxy, excluding the common variants, synonymous variants and non-synonymous variants that are predicted to have no deleterious effect on protein function.
Thirty-four potential disease-causing heterozygous variants were identified in 11 of 37 candidate genes, including _ARMC4_, _CCDC40_, _CCDC65_, _DHAH8_, _DNAAF2_, _DNAH11_, _DNAH5_, _DNAH8_,
_DRC1_, _HYDIN_, and _SPAG1_ (Table 2). These mutated genes were distributed in 19 CHD/HTX patients (45.2%, 19/42). Eighteen CHD patients with heterotaxy and CD had 31 variants in 8 genes,
and 1 CHD patient (patient #5030) with heterotaxy and normal ciliary function possessed 3 variants in 3 genes. These findings indicate that the CHD patients with heterotaxy and CD has a
significantly higher gene mutation rate than the CHD patients with heterotaxy and normal ciliary function among the CHD patients with heterotaxy (62.1%, 18/29 vs. 7.7%, 1/13; _P_ = 0.0018)
(Table 3). ASSOCIATION OF _DNAH11_ VARIANTS WITH THE RISK OF CHD AND HETEROTAXY SYNDROME As shown in Table 4, the _DNAH11_ gene showed the highest mutation rate (16.7%; 14 of 84 alleles)
among the CHD patients with heterotaxy. The _HYDIN_ and _DNAH8_ genes both showed 6% mutation rate (5 of 84 alleles). The mutation rate of _DNAH5_ genes was 4.8% (4 of 84 alleles), and the
other genes (_ARMC4_, _CCDC40_, _CCDC65_, _DHAH8_, _DNAAF2_, _DRC1_, and _SPAG1_) all showed a 1.2% mutation rate (1 of 84 alleles). Concerning the highest mutation rate in the _DNAH11_
gene, we further analyzed the association of _DNAH11_ mutations with CHD and heterotaxy syndrome. In this study, there were 14 mutations in the _DNAH11_ genes in 7 of 42 patients, including
11 missense, 1 frameshift and 2 stop-gain mutations. Eight of these variants were novel and not present in the ExAC or 1000 Genomes Project databases, and 6 were low-frequency variants (MAF
< 0.01%). Conservation analysis was processed via UCSC Genome Browser hg19. In our previous study, whole genome sequencing was performed in 98 CHD patients without heterotaxy, and the
methodology used in the previous study is strictly the same as that used in the present investigation. The CHD subtypes of these 98 CHD patients are listed in Table 5. By consulting the
previous exome database in our laboratory, we found no disease-causing mutations in the _DNAH11_ gene among 98 CHD cases. Moreover, one _DNAH11_ mutation (c.A9584G:p.N3195S) was found in 1
case (Patient 2073) among 3 CHD patients with heterotaxy. Combined with the results showing 7 of 42 patients with _DNAH11_ mutations in this study, the prevalence of _DNAH11_ mutations was
higher in CHD patients with heterotaxy (8 of 45 cases) than in CHD patients (0 of 98 cases). The 98 CHD cases were considered controls because these cases exhibited only CHD, and the
association of _DNAH11_ mutations with CHD and heterotaxy was significant (8 of 45 CHD patients with heterotaxy vs. 0 of 98 controls, _P_ = 0.0001 by Fisher’s exact test). These findings
suggest a significant association of _DNAH11_ mutations with the risk of CHD and heterotaxy syndrome (Table 6). _DNAH11_ COMPOUND HETEROZYGOUS MUTATIONS IN CHD FAMILIES WITH HETEROTAXY
Disease-causing _DNAH11_ mutations are inherited by autosomal recessive inheritance. Because there were no homozygous mutations in _DNAH11_ in this study, we focused on compound heterozygous
mutations in the _DNAH11_ gene, which may be the main cause of the development of CHD/heterotaxy. The 14 disease-causing heterozygous mutations in _DNAH11_ were distributed among 7 CHD
patients with heterotaxy, with 6 patients having two or more _DNAH11_ mutations. These _DNAH11_ mutations were further confirmed to be present in the available DNA of parents and other
family members of the patients by Sanger sequencing. Interestingly, six different compound heterozygous variants in _DNAH11_ were validated respectively in six different families, including
family 1-5031 (p.W802X/p.M282I), family 2-5045 (p.T3460K/p.G4425S), family 3-5065 (p.G447R/p.L1157R), family 4-5130 (p.I2262T/p.D3800H), family 5-5707 (p.S1823fs/p.F2759L/p.R4395X) and
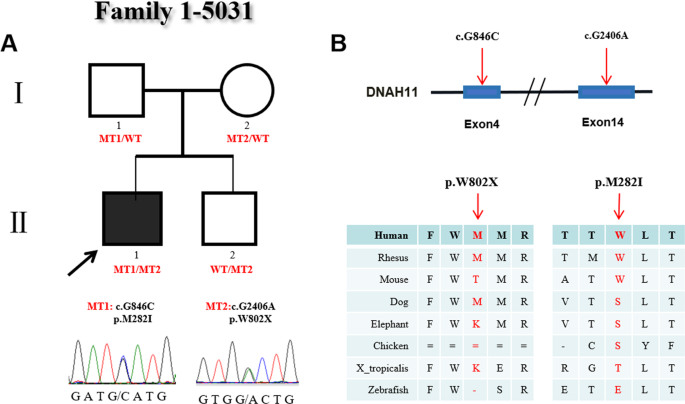
family 6-5062 (p.D3610V/p.I243V) (Table 7). In family 1 (Fig. 1A), there were four members. The proband (#5031), who carried 2 heterozygous mutations (c.G2406A:p.W802X and c.G846C:p.M282I),
was male and diagnosed with CHD and heterotaxy, including isolated right heart, complete atrioventricular canal (CAVC), double outlet right ventricle (DORV) and atrial septal defect (ASD),
and showed abnormal ciliary function. His parents and younger brother were all without clinical manifestations, but their ciliary movements were abnormal as evidenced by uncoordinated
ciliary waves. In this family, the heterozygous variant c.G2406A (p.W802X) was a _novo_ stop-gain mutation located on exon 14 of _DNAH11_. This variant was present in the proband and his
young brother and inherited from their mother. Functional analysis showed the variant p.W802X was predicted to be damaging by SIFT software and conserved among human, rhesus and dog species.
The other heterozygous variant c.G846C (p.M282I) located in exon 4 in the proband was transmitted from his father and was not observed in his young brother and mother. This mutation was
reported in the Exome Aggregation Consortium (ExAc) (0.00006547) and 1000 Genomes Project (0.000199681) databases. The p.M282I change was also predicted to be damaging by SIFT software and
conserved among human, rhesus and mouse species (Fig. 1B). In family 2, the proband (#5045) was male and diagnosed with CHD and heterotaxy, showing abnormal ciliary movement. His parents
were both heterozygous carriers and showed normal phenotypes. The heterozygous variants (c.C10379A:p.T3460K/c.G13273A:p.G4425S) of the _DNAH11_ gene were confirmed in family 2 by Sanger
sequencing (Fig. 2A). Of the two heterozygous variants, c.C10379A (p.T3460K) was a missense variant type located in exon 64 and was inherited from the proband’s mother and presented in the
ExAc (0.0001879) and 1000 Genomes Project (0.000199681) databases. The amino acid p.T3460K alteration is predicted to be damaging by SIFT, PolyPhen2 and MutationTaster and is highly
conserved among mammalian species but not in chickens. The other variant, c.G13273A (p.G4425S), was a novel missense variant type located in exon 81 and which was transmitted from the
proband’s carrier father. The p.G4425S change was predicted to be damaging by the SIFT, PolyPhen2 and MutationTaster software programs and is highly conserved among mammalian species (Fig.
2B). The family 3 proband (#5065) had severe CHD and heterotaxy, with abnormal ciliary movement. His parents were normal and healthy. This family carried two heterozygous missense mutations
(c.G1339A:p.G447R/c.T3470G:p.L1157R) in the proband (Fig. 3A). The c.G1339A (p.G447R) in exon 7 was a novel variant and absent in his parents, indicating that this variant was _de novo_. The
altered amino acid p.G447R was conserved among the human, rhesus, dog and elephant species. Functional analysis of this change was predicted to be benign by prediction software. The other
c.T3470G (p.L1157R) in exon 18 was a missense mutation type and presented in the ExAc (0.000149) and 1000 Genomes Project (0.000199) databases. The mutation c.T3470G was inherited from his
mother, resulting in the substitution of the 1157 amino acid Leu (L) with Arg (R) (p.L1157R) and was predicted to be a damaging change in the protein. The altered amino acid p.L1157R is
highly conserved among many species. Although the p.G447R variant was predicted to be not damaging, considering the proband’s mother normal phenotype, we concluded that the combined effects
of the two heterozygous variants (p.G447R/p.L1157R) may be an important factor causing CHD/heterotaxy disease (Fig. 3B). The family 4 proband (#5130), who was female and diagnosed with CHD,
heterotaxy and CD, carried two heterozygous missense variants (c.T6785C:p.I2262T/c.G11398C:p.D3800H). We obtained the proband and her mother’s blood sample for validating the detected
variants by Sanger sequencing. The blood sample of the proband’s father was not obtained. However, the parents both had normal phenotypes according to the medical history record (Fig. 4A).
The c.T6785C (p.I2262T) variant was a novel heterozygous type and was located in exon 41 of _DNAH11_, which was inherited from the proband’s mother. Functional analysis indicated that the
p.I2262T variant was predicted to be damaging by the SIFT, PolyPhen2 and Mutation Taster software programs and was highly conserved among many species. The variant c.G11398C (p.D3800H) in
exon 70 was also a _novel_ heterozygous variant and was absent from the ExAc and 1000 Genomes Project databases. Functional analysis predicted that the variant p.D3800H was damaging (by
MutationTaster software) and was conserved among different species (Fig. 4B). We could not collect a blood sample from the proband’s father; therefore the inherited origin of the c.G11398C
(p.D3800H) variant could not be determined, and we proposed that the compound heterozygous variants (p.I2262T/p.D3800H) may be associated with the development of the proband’s disease. In
family 5, patient #5707, diagnosed with CHD, heterotaxy and CD, had three variants in _DNAH11_, one frameshift variant (c.5470dupC:p.S1823fs), one missense variant (c.T8275C:p.F2759L) and
one stop-gain variant (C13183T:p.R4395X). We did not obtain a blood sample from the patient’s parents, so we only confirmed these variants in the DNA sample of patient #5707 by Sanger
sequencing. One frameshift variant (c.5470dupC:p.S1823fs) and two variants (c.T8275C:p.F2759L and C13183T:p.R4395X) were validated (Fig. 5A). The variant (c.5470dupC:p.S1823fs) was a _novel_
frameshift insertion located in exon 32 and affected the protein function, which was conserved among different species. The c.T8275C variant was a missense variant located in exon 50 and
was presented in the ExAc (0.0003) and 1000 Genomes Project (0.000599042) databases. The p.F2759L change was predicted to be damaging by the PolyPhen2 and MutationTaster software programs
and was conserved among different species. The variant (C13183T:p.R4395X) in exon 81 was a _novel_ stop-gain type that influenced protein function. The p.R4395X variant was highly conserved
among different species (Fig. 5B). In family 6 (Fig. 6A), there were three members. The proband (#5062), who carried 2 heterozygous mutations (c.A10829T:p.D3610V and c.A727G:p.I243V), was
female and diagnosed with CHD and heterotaxy, including isolated right heart, PA, levo-transposition of the great arteries (L-TGA) and ASD, and showed abnormal ciliary function. His parents
both showed normal phenotypes. The c.A10829T (p.D3610V) variant was a _novel_ heterozygous type located in exon 66 of _DNAH11_ and was inherited from the proband’s mother. Functional
analysis showed the variant p.D3610V was predicted to be damaging by SIFT software and conserved among human, rhesus and dog species. The other heterozygous variant c.A727G (p.I243V) was a
_novel_ missense mutation type located in exon 4 of _DNAH11_ and was transmitted from the proband’s father. The p.I243V change was also predicted to be damaging by SIFT software and
conserved among human, rhesus and mouse species (Fig. 6B). Based on these confirmed variants and the medical history of the families, the findings suggest that these _DNAH11_ compound
heterozygote variants are responsible for the development of CHD/heterotaxy syndrome. DISCUSSION Human LR asymmetry plays an important role in normal organogenesis and provides the
developmental basis for correct heart looping2. LR asymmetry disorders in early embryonic development may result in a series of congenital birth defects, such as heterotaxy syndrome14.
Although many genes have been reported to be associated with LR asymmetry disorders15, the exact aetiological mechanism of CHD and heterotaxy remain unknown. Moreover, studies have found
that cilia participate in the formation of the left and right asymmetric mode by regular swinging to form a nodal flow during the embryonic period16. PCD and CD have been shown to be
associated with CHD and heterotaxy syndrome. PCD is considered a monogenic heterogeneous recessive disorder, and CHD and heterotaxy syndrome are multiple, complex inherited diseases. There
may be a link between PCD and CHD/HTX. In this study, we found 34 potential disease-causing heterozygous variants in 11 genes present in 19 CHD/HTX patients, accounting for 45.2% of 42
CHD/HTX patients. We compared the gene mutation rate in the CHD/HTX patients with CD and CHD/HTX patients without CD and found that CHD/HTX patients with CD had a significantly higher gene
mutation rate than CHD/HTX patients without CD. The results suggest that PCD-related gene mutations are significantly associated with CHD with heterotaxy and CD. _DNAH11_ is localized to the
proximal region of respiratory cilia and is known as a PCD-related gene. Approximately 6% of PCDs are caused by _DNAH11_ mutations17. Although cilia are required for LR body-axis
determination and second heart field (SHF) Hedgehog (Hh) signalling, Burnicka _et al_. observed that _DNAH11_ mutations did not disrupt SHF Hh signalling and caused AVSDs only concurrently
with heterotaxy, a LR axis abnormality18. _DNAH11_ showed the highest mutation rate in this study, followed by _HYDIN_ and _DNAH5_. We concluded that _DNAH11_ mutations may be an important
risk factor involved in the development of CHD/HTX syndrome. We reanalyzed the exome database in our previous study, including 98 CHD cases and 3 CHD/HTX cases, and found no mutation in the
_DNAH11_ gene among 98 CHD cases and 1 _DNAH11_ mutation in 1 of 3 CHD patients with heterotaxy. We considered the 98 CHD cases as controls, fisher’s exact test revealed that _DNAH11_
mutations can significantly increase the risk of developing CHD/HTX syndrome, indicating a significant association of _DNAH11_ mutations with CHD and heterotaxy syndrome. It is known that
_DNAH11_ mutations are inherited by autosomal recessive inheritance pattern. Bartoloni L _et al_. found a homozygous nonsense mutation (R2852X) in _DNAH11_ in a patient with situs inversus
totalis19. _DNAH11_ compound heterozygotes (p.R2250*/p.Q3604*) were observed in two monochorionic biamniotic male twins with PCD13. Nader Nakhleh _et al_. found _DNAH11_ compound
heterozygotes (p.Q1507P/p.E3133K) in a patient with heterotaxy; these two mutations were predicted to be damaging and involved in hyperkinetic ciliary beats4,20. _DNAH11_ homozygous
mutations were not observed in our study. We focused on recessive _DNAH11_ mutations in the families and found six different compound heterozygous variants in six families and concluded that
these compound heterozygous variants may be the main factors causing CHD/heterotaxy syndrome. Recently, a study found that heterotaxy patients with heterozygous _DNAH6_ mutations also had
heterozygous mutations in _DNAH5_ and _DNAHI1_ genes, which experimentally showed that the trans-heterozygous interactions of _DNAH6_ with _DNAI1_ or _DNAH5_ may contribute to heterotaxy
syndrome21. In our study, in addition to these _DNAH11_ compound heterozygotes, we also found that the patients with the _DNAH11_ heterozygous mutations possessed other heterozygous
mutations in known PCD genes, such as _HYDIN_ and _DNAH5_. Patients 5033, who possessed two different heterozygous variants in the _DNAH11_ and _HYDIN_ genes (Table 2), presented primary CD
and heterotaxy/CHD, indicating that interactions between trans-heterozygous variants of _DNAH11_ and _HYDIN_ may be involved in heterotaxy/CHD and PCD. Although over 40 PCD pathogenic genes
were revealed, our study found compound heterozygous variants in only _DNAH11_ among the CHD patients with heterotaxy, which is subject to the limited number of patients recruited in this
study. We speculated that larger studies may uncover comprehensive genetic pathogenic factors related to cilia among these patient populations. However, we can still conclude that pathogenic
_DNAH11_ mutations are an important cause of heterotaxy with CHD. Additionally, we did not conduct deeper functional studies of these _DNAH11_ heterozygous variants for pathogenic
confirmation and further interpretation of the genotypic and phenotypic mechanisms, which should be the key aspects of future works. CONCLUSION In summary, we performed targeted sequencing
and Sanger sequencing to analyze the exonic regions of 37 candidate genes in 42 HTX patients with CHD from unrelated families and found 34 potential disease-causing heterozygous variants in
11 genes among the 19 CHD patients with heterotaxy syndrome. The association of _DNAH11_ variants with CHD and heterotaxy was significant (_P_ = 0.0001). In families, six different compound
heterozygous variants of _DNAH11_ were validated in family 1-5031 (p.W802X/p.M282I), family 2-5045 (p.T3460K/p.G4425S), family 3-5065 (p.G447R/p.L1157R), family 4-5130 (p.I2262T/p.D3800H),
family 5-5707 (p.S1823fs/p.F2759L/p.R4395X) and family 6-5062 (p.D3610V/p.I243V). These findings expand the spectrum of _DNAH11_ gene mutations causing the development of HTX/CHD and provide
an important clue for understanding the genetic mechanism of HTX/CHD syndrome. METHODS PATIENT COHORTS Blood samples from patients or their family members were collected in accordance with
the Declaration of Helsinki, and the Ethics Committees of Children’s Hospital of Fudan University (CHFU) approved this study. Written informed consent was obtained from the parents and
guardians of all the probands. Informed consent has been obtained for the blood samples taken from the available family members. Forty-two participants with CHD and heterotaxy syndrome were
recruited from the CHFU, Shanghai, China, including 28 males and 14 females, with ages ranging from 0.1 to 12.1 years. Family medical history was obtained at the cardiovascular centre of the
CHFU, and medical records were reviewed to confirm the disease diagnosis. Blood samples from probands and their available family members were obtained for further genetic analysis. NASAL
TISSUE SAMPLING AND CILIARY MOTION ANALYSIS Nasal tissues were collected from patients with Rhino-Probe (Arlington Scientific, Springville, UT) curettage of the inferior nasal turbinate.
Exclusion criteria included severe bleeding diathesis or conditions such as haemophilia or hereditary haemorrhagic telangiectasia syndrome. The nasal tissues were suspended in L-15 medium
(Invitrogen, CA) for videomicroscopy using a Leica inverted microscope (DMIRE2) with a 67× oil objective under differential interference contrast optics. Movies were recorded at 200 frames/s
at room temperature using a 680 PROSILICA GE camera (Allied Vision, PA), and digital recordings were evaluated by a blinded panel of coinvestigators. Abnormal ciliary motion was described
as follows: immotile (I), discordance (D), wave (W), restricted (R) and no cilia (None). BLOOD DNA EXTRACTION A QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) was used to extract blood
genomic DNA from probands and available family members according to the manufacturer’s instructions. The concentration and purity of genomic DNA were measured by absorbance at 260 and 280 nm
by using a NanoDropTM 1000 Spectrophotometer (Thermo Scientific, Wilmington, USA). The extracted genomic DNA were stored at −80 °C until the samples were ready for further analysis.
TARGETED SEQUENCING ANALYSIS OF GENE VARIANTS Targeted sequencing analysis was performed for the 42 probands from the CHFU. We focused on 37 candidate PCD-related genes, namely, _ABCC4_,
_ARMC4_, _C21orf59_, _CCDC39_, _CCDC40_, _CCDC65_, _CCDC114_, _CCDC151_, _CCNO_, _DNAAF1_, _DNAAF2_, _DNAFF3_, _DNAAF5_, _DNAH5_, _DNAH8_, _DNAH11_, _DNAI1_, _DNAI2_, _DNAL1_, _DRC1_,
_DYX1C1_, _HEATR2_, _HYDIN_, _LRRC6_, _NAT10_, _NME8_, _PTGES_, _PTGES2_, _PTGES3_, _PTGER4_, _PTGS1_, _PTGS2_, _RSPH1_, _RSPH4A_, _RSPH9_, _SPAG1_ and _ZMYND10_, intending to find novel or
rare coding variants present in patients with CHD and heterotaxy syndrome. Primers covering all exons and at least 10 bp of all intron/splice sites of these genes were designed online
(https://www.ampliseq.com/). The libraries were constructed using the Ion AmpliSeq Library Kit v2.0 (Life Technologies, USA) according to the protocol. The concentration of each library was
confirmed by a TaqMan Quantification Kit (Life Technologies). The Ion OneTouch 2 system with an Ion PGM Template OT2 200 Kit (Life Technologies, USA) was used to amplify pooled barcode
libraries, and ion sphere particles (ISP) were enriched according to the E/S module protocol. The enriched template-positive ISPs were loaded and sequenced on an Ion 316™ Chip by PGM (Life
Technologies, USA). For each subject, base calls were detected with Torrent Suite software. Raw sequencing data were aligned against the human reference genome GRCh37/hg19 (NCBI) using
NextGENe software. Single-nucleotide variations (SNVs) were aligned based on the following criteria: 1) the variant was detected on both strands of the sequence reads; 2) the minimum
coverage of reads was no less than 10×; 3) the variant reads represented more than 20% of the sequence reads. The variants filtered from NextGENe software were confirmed by Sanger sequencing
and compared with 1000 Genomes (http://www.1000genomes.org) and ExAc databases (http://exac.broadinstitute.org/) as well as our laboratory’s internal databases. Additionally, the risk of
SNVs was predicted using the silico tools SIFT (http://sift.jcvi.org/), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (http://www.mutationtaster.org/). The amino acid
changes were compared among eight mammalian species by conservation analysis (UCSC Genome Browser hg19). STATISTICAL ANALYSIS Statistical analysis was performed using the Chi-square test
with GraphPad Prism 6.0 Software. Fisher’s exact test was used to analyze the association of _DNAH11_ mutations with CHD and heterotaxy. Values were considered significant at _P_ < 0.05.
DATA AVAILABILITY The datasets generated during and/or analysed in this study are available from the corresponding author on reasonable request. REFERENCES * van der Linde, D. _et al_. Birth
Prevalence of Congenital Heart Disease Worldwide A Systematic Review and Meta-Analysis. _Journal of the American College of Cardiology_ 58, 2241–2247,
https://doi.org/10.1016/j.jacc.2011.08.025 (2011). Article PubMed Google Scholar * Deng, H., Xia, H. & Deng, S. Genetic basis of human left-right asymmetry disorders. _Expert Rev Mol
Med_ 16, e19, https://doi.org/10.1017/erm.2014.22 (2015). Article CAS PubMed Google Scholar * Lin, A. E., Ticho, B. S., Houde, K., Westgate, M. N. & Holmes, L. B. Heterotaxy:
associated conditions and hospital-based prevalence in newborns. _Genet Med_ 2, 157–172, https://doi.org/10.1097/00125817-200005000-00002 (2000). Article CAS PubMed Google Scholar *
Nakhleh, N. _et al_. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. _Circulation_ 125, 2232–2242,
https://doi.org/10.1161/CIRCULATIONAHA.111.079780 (2012). Article PubMed PubMed Central Google Scholar * Harrison, M. J., Shapiro, A. J. & Kennedy, M. P. Congenital Heart Disease and
Primary CiliaryDyskinesia. _PaediatrRespirRev_ 18, 25–32, https://doi.org/10.1016/j.prrv.2015.09.003 (2016). Article Google Scholar * Klena, N. T., Gibbs, B. C. & Lo, C. W. Cilia and
Ciliopathies in Congenital Heart Disease. _Cold Spring Harb Perspect Biol_ 9, https://doi.org/cshperspect.a028266 (2017). * Harden, B. _et al_. Increased postoperative respiratory
complications in heterotaxy congenital heart disease patients with respiratory ciliary dysfunction. _J Thorac CardiovascSurg_ 147, 1291–1298 e1292,
https://doi.org/10.1016/j.jtcvs.2013.06.018 (2014). Article Google Scholar * Lin, A. E. _et al_. Laterality defects in the national birth defects prevention study (1998–2007): birth
prevalence and descriptive epidemiology. _Am J Med Genet A_ 164A, 2581–2591, https://doi.org/10.1002/ajmg.a.36695 (2014). Article PubMed Google Scholar * Sutherland, M. J. & Ware, S.
M. Disorders of left-right asymmetry: heterotaxy and situs inversus. _Am J Med Genet C Semin Med Genet_ 151C, 307–317, https://doi.org/10.1002/ajmg.c.30228 (2009). Article CAS PubMed
Google Scholar * Li, Y. _et al_. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. _Nature_ 521, 520–524, https://doi.org/10.1038/nature14269
(2015). Article ADS CAS PubMed PubMed Central Google Scholar * Chapelin, C. _et al_. Isolation of several human axonemal dynein heavy chain genes: genomic structure of the catalytic
site, phylogenetic analysis and chromosomal assignment. _Febs Letters_ 412, 325–330, https://doi.org/10.1016/S0014-5793(97)00800-4 (1997). Article CAS PubMed Google Scholar * Dougherty,
G. W. _et al_. DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes. _Am J Respir Cell Mol Biol_ 55, 213–224,
https://doi.org/10.1165/rcmb.2015-0353OC (2016). Article CAS PubMed PubMed Central Google Scholar * Lai, M. _et al_. Gene editing of DNAH11 restores normal cilia motility in primary
ciliary dyskinesia. _J Med Genet_ 53, 242–249, https://doi.org/10.1136/jmedgenet-2015-103539 (2016). Article CAS PubMed Google Scholar * Mortari, E. P. _et al_. Heterotaxy syndrome with
and without spleen: Different infection risk and management. _Journal of Allergy and Clinical Immunology_ 139, 1981–1984.e1, https://doi.org/10.1016/j.jaci.2016.10.014 (2017). Article
Google Scholar * Brueckner, M. Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. _Circulation_ 115, 2793–2795, https://doi.org/10.1161/CIRCULATIONAHA.107.699256 (2007).
Article PubMed Google Scholar * Wang, G. L., Yost, H. J. & Amack, J. D. Analysis of Gene Function and Visualization of Cilia-Generated Fluid Flow in Kupffer’s Vesicle. _Jove-Journal
of Visualized Experiments_, https://doi.org/10.3791/50038 (2013). * Pifferi, M. _et al_. New DNAH11 mutations in primary ciliary dyskinesia with normal axonemal ultrastructure. _European
Respiratory Journal_ 35, 1413–1416, https://doi.org/10.1165/rcmb.2015-0353OC (2010). Article CAS PubMed Google Scholar * Burnicka-Turek, O. _et al_. Cilia gene mutations cause
atrioventricular septal defects by multiple mechanisms. _Human Molecular Genetics_ 25, 3011–3028, https://doi.org/10.1093/hmg/ddw155 (2016). Article CAS PubMed PubMed Central Google
Scholar * Bartoloni, L. _et al_. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia.
_Proceedings of the National Academy of Sciences of the United States of America_ 99, 10282–10286, https://doi.org/10.1073/pnas.152337699 (2002). Article ADS CAS PubMed PubMed Central
Google Scholar * Nakhleh, N. _et al_. High Prevalence of Respiratory Ciliary Dysfunction in Congenital Heart Disease Patients With Heterotaxy. _Circulation_ 125, 2232–U2164,
https://doi.org/10.1161/CIRCULATIONAHA.111.079780 (2012). Article PubMed PubMed Central Google Scholar * Li, Y. _et al_. DNAH6 and Its Interactions with PCD Genes in Heterotaxy and
Primary CiliaryDyskinesia. _PloSGenet_ 12, e1005821, https://doi.org/10.1371/journal.pgen.1005821 (2016). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was
supported by grants from the National Key Research and Development Program of China (2016YFC1000500) and the Natural Science Foundation of China (81570282, 81370198 and 81873482) and Science
and Research Foundation of Shanghai Municipal Commission of Health and Family Planning for Young Scientists (20144Y0057). AUTHOR INFORMATION Author notes * Sida Liu and Weicheng Chen
contributed equally. AUTHORS AND AFFILIATIONS * Children’s Hospital of Fudan University, Shanghai, 201102, China Sida Liu, Weicheng Chen, Yongkun Zhan, Shuolin Li, Xiaojing Ma, Wei Sheng
& Guoying Huang * Shanghai Key Laboratory of Birth Defects, Shanghai, 201102, China Xiaojing Ma, Duan Ma, Wei Sheng & Guoying Huang * Key Laboratory of Molecular Medicine, Ministry
of Education, Shanghai Medical College, Fudan University, Shanghai, 200000, China Duan Ma Authors * Sida Liu View author publications You can also search for this author inPubMed Google
Scholar * Weicheng Chen View author publications You can also search for this author inPubMed Google Scholar * Yongkun Zhan View author publications You can also search for this author
inPubMed Google Scholar * Shuolin Li View author publications You can also search for this author inPubMed Google Scholar * Xiaojing Ma View author publications You can also search for this
author inPubMed Google Scholar * Duan Ma View author publications You can also search for this author inPubMed Google Scholar * Wei Sheng View author publications You can also search for
this author inPubMed Google Scholar * Guoying Huang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Conception and study design: Guoying
Huang and Wei Sheng. Acquisition of patient information and communication with the patients’ families: Sida Liu, Weicheng Chen and Shuolin Li. NGS design, Sanger sequencing and data
analysis: Sida Liu, Yongkun Zhan, Shuolin Li, Duan Ma and Xiaojing Ma. Drafting: Sida Liu. Manuscript editing and revision: Guoying Huang, Wei Sheng and Sida Liu. CORRESPONDING AUTHORS
Correspondence to Wei Sheng or Guoying Huang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Liu, S., Chen, W., Zhan, Y. _et al._ _DNAH11_ variants and its association with
congenital heart disease and heterotaxy syndrome. _Sci Rep_ 9, 6683 (2019). https://doi.org/10.1038/s41598-019-43109-6 Download citation * Received: 27 November 2018 * Accepted: 16 April
2019 * Published: 30 April 2019 * DOI: https://doi.org/10.1038/s41598-019-43109-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative