Gemini quaternary ammonium compound pmt12-bf4 inhibits candida albicans via regulating iron homeostasis
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Quaternary ammonium compounds (QACs) are classified as cationic surfactants, and are known for their biocidal activity. However, their modes of action are thus far not completely
understood. In this study, we synthesized a gemini QAC, PMT12-BF4 and found that it exerted unsurpassed broad-spectrum antifungal activity against drug susceptible and resistant _Candida
albicans_, and other pathogenic fungi, with a minimal inhibitory concentration (MIC) at 1 or 2 μg/mL. These results indicated that PMT12-BF4 used a mode of action distinct from current
antifungal drugs. In addition, fungal pathogens treated with PMT12-BF4 were not able to grow on fresh YPD agar plates, indicating that the effect of PMT12-BF4 was fungicidal, and the minimal
fungicidal concentration (MFC) against _C. albicans_ isolates was 1 or 2 μg/mL. The ability of yeast-to-hyphal transition and biofilm formation of _C. albicans_ was disrupted by PMT12-BF4.
To investigate the modes of action of PMT12-BF4 in _C. albicans_, we used an RNA sequencing approach and screened a _C. albicans_ deletion mutant library to identify potential pathways
affected by PMT12-BF4. Combining these two approaches with a spotting assay, we showed that the ability of PMT12-BF4 to inhibit _C. albicans_ is potentially linked to iron ion homeostasis.
SIMILAR CONTENT BEING VIEWED BY OTHERS 3-HYDROXY COUMARIN DEMONSTRATES ANTI-BIOFILM AND ANTI-HYPHAL EFFICACY AGAINST _CANDIDA ALBICANS_ VIA INHIBITION OF CELL-ADHESION, MORPHOGENESIS, AND
VIRULENT GENES REGULATION Article Open access 19 July 2023 VITAMIN B5 METABOLISM IS ESSENTIAL FOR VACUOLAR AND MITOCHONDRIAL FUNCTIONS AND DRUG DETOXIFICATION IN FUNGI Article Open access 23
July 2024 FUNGAL CELL BARRIERS AND ORGANELLES ARE DISRUPTED BY POLYHEXAMETHYLENE BIGUANIDE (PHMB) Article Open access 16 February 2023 INTRODUCTION _Candida albicans_ is an opportunistic
fungal pathogen causing candidiasis mainly in immunocompromised individuals with candidemia, or superficial infections such as oral thrush and vaginal infections1. To date, polyenes, azoles,
and echinocandins are the dominant antifungal classes used to cure candidiasis1. However, increased incidence of drug resistance has reduced the efficacy of drugs in these classes,
indicating an urgent need to develop antifungal agents to combat drug-resistant _C. albicans_. Simple surfactants whose structures are based on quaternary ammonium compounds (QACs) are
usually made of a positive nitrogen atom with four substituents attached to the nitrogen atom, at least one of which is a long alkyl chain. QACs have many applications, for example in
anesthesiology, dentistry, ophthalmology, and asthma2,3,4,5. A single range of applications is directly related to their biocidal activity. QACs have biocidal activity not only against
Gram-positive and Gram-negative bacteria but also against fungal pathogens6,7. However, there are limited studies on gemini surfactants derived from QACs. Gemini surfactants are made of two
polar heads separated by a spacer. Polar heads can have a positive charge when synthesized from quaternary ammonium salts, and they can be substituted with alkyl chains called tails. With
the presence of hydrophobic rigid or flexible spacers and two identical or different hydrophilic heads, it becomes possible to synthesize dimeric surfactants with diverse structures8. Gemini
surfactants have significantly higher surface activity compared to conventional analogues. The reason for the increased activity of gemini surfactants is the larger total number of carbon
atoms in the hydrophobic chains9,10. Moreover, their biocidal properties are noteworthy and characterized by a broad spectrum of antimicrobial activity11,12. However, the potential
mechanisms that gemini QACs use to target pathogens remains unknown. In this study, we demonstrated that a newly synthesized gemini QAC, 1, 5-bis(dodecyl)−1, 1, 3, 5, 5-pentamethyl-3-aza-1,
5-pentanediammonium ditetrafluoroborate (PMT12-BF4), possesses novel fungicidal activity against a broad spectrum of fungal pathogens including drug-resistant _C. albicans_. The hyphal
growth and biofilm formation of _C. albicans_ were reduced after PMT12-BF4 treatment, and the mode of action of this compound was potentially associated with iron ion homeostasis based on
RNA sequencing, mutant library screening, and spotting assays. RESULTS SYNTHESIZED SURFACTANTS EXHIBITED A BROAD SPECTRUM OF ANTIFUNGAL ACTIVITY Two gemini QACs (PMT12-BF4 and PMT16-BF4)
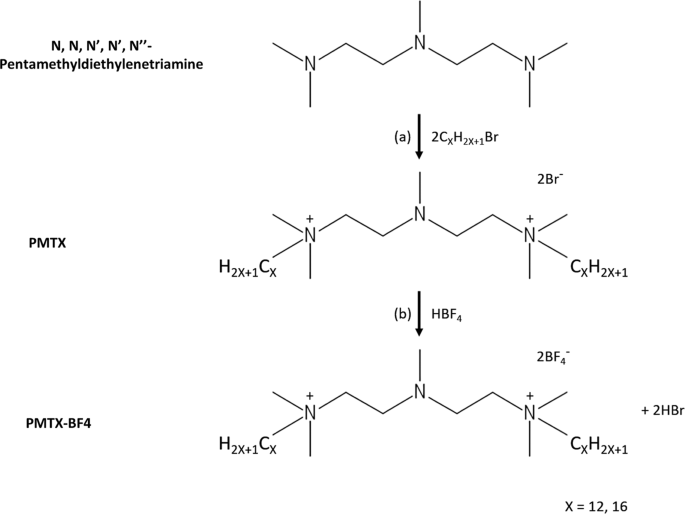
were synthesized (Fig. 1) to test antifungal activity against clinical drug-susceptible and -resistant _C. albicans_ isolates, and multiple fungal pathogens (Table 1). PMT12-BF4 with 12
carbons in the alkyl chain exhibited novel antifungal activity against _C. albicans_ SC5314 with MIC = 1 μg/mL, as well as drug-resistant 12–99 and 89 isolates with both MIC = 1 μg/mL (Table
2). In addition, PMT12-BF4 was also effective against non-_albicans Candida_ species including _C. tropicalis_ MYA3404 (2 μg/mL) and _C. glabrata_ CBS138 (1 μg/mL), and other human or plant
pathogenic fungi such as _Cryptococcus neoformans_ H99 (1 μg/mL)_, Aspergillus fumigatus_ AF293 (2 μg/mL), _Fusarium oxysporum_ FOSC3-a (2 μg/mL), and _Fusarium oxysporum_ f. sp.
_lycopersici_ 4287 (2 μg/mL) (Table 2). In contrast, the analogue PMT16-BF4 which possessed an alkyl chain with 16 carbons was not effective against _C. albicans_ (MIC > 64 μg/mL) (Table
2), indicating the critical role of the length of the alkyl chain in gemini QACs. Interestingly, not all fungal pathogens tested showed similar susceptibility to PMT16-BF4. _C. glabrata_
CBS138 and _C. neoformans_ H99 showed higher susceptibility (MIC = 4 μg/mL) to this compound. Moreover, growth of _F. oxysporum_ f. sp. _lycopersici_ 4287, _F. oxysporum_ and _A. fumigatus_
AF293 was also inhibited by PMT16-BF4. Interestingly, the dandruff borne fungus, _Malassezia furfur_, was resistant to both PMT12-BF4 and PMT16-BF4 (_i.e_., MIC > 64 μg/mL), indicating
the compound’s target(s) in _M. furfur_ might be distinct from other compound-susceptible fungi (Table 2). PMT12-BF4 EXHIBITED FUNGICIDAL ACTIVITY AGAINST _C. ALBICANS_ To determine whether
these gemini QACs were fungicidal, MFCs were obtained by spotting assay after MIC determination. We demonstrated that PMT12-BF4 was fungicidal to all fungal pathogens tested except _M.
furfur_, while PMT16-BF4 was only fungicidal to _C. glabrata_, _C. neoformans_ and _F. oxysporum_ FOSC3-a (Table 2). Furthermore, as shown in the growth kinetics assay, _C. albicans_ cells
treated with the MIC of PMT12-BF4 in YPD (2 μg/mL) did not increase even after incubation for 48 h (Fig. 2A). The fungicidal activity of PMT12-BF4 against _C. albicans_ was further confirmed
by time-killing assays. The number of colony forming units (CFUs) decreased >99% compared to initial inoculum after 48 h incubation at 4 μg/mL PMT12-BF4 (Fig. 2B). PMT12-BF4 REDUCED
YEAST-TO-HYPHAE TRANSITION IN _C. ALBICANS_ The ability to undergo yeast-to-hyphae transition is a critical virulence factor in _C. albicans_. To determine this ability in _C. albicans_,
cultures were grown overnight in YPD, washed twice with ddH2O, and incubated at 37 °C with RPMI 1640 medium to induce hyphal development. The length of germ tubes decreased after 3 h
incubation in the presence of 0.25 μg/mL PMT12-BF4, indicating that PMT12-BF4 can interfere with morphological transition in _C. albicans_ (Fig. 3A). Moreover, cells exposed to PMT12-BF4 at
2 μg/mL showed wrinkled cell surfaces and even broken cells under SEM observation. (Fig. 3B). PMT12-BF4 INTERFERED WITH _C. ALBICANS_ BIOFILM FORMATION The ability of _C. albicans_ to form a
biofilm is usually linked to drug tolerance. We thus tested PMT12-BF4 interference of biofilm formation in _C. albicans_. There was reduction in biofilm formation after addition of
PMT12-BF4 at 2 or 4 μg/mL in drug-susceptible strain SC5314 and echinocandin-resistant isolate 89, and the biofilm structure could be easily removed by gently pipetting up and down during
the wash step (Fig. 4A). The fluconazole-resistant isolate 12–99 only showed reduced biofilm formation at 4 μg/mL PMT12-BF4 (Fig. 4A). In summary, the biofilm formation was decreased by more
than 50% with treatment with PMT12-BF4 at 4 μg/mL in three _C. albicans_ strains (SC5314, 12–99, and 89) as compared with the control group (Fig. 4B). GENOME-WIDE ANALYSIS OF
PMT12-BF4-MEDIATED GENES IN _C. ALBICANS_ To investigate PMT12-BF4-mediated genes, we performed transcriptome analysis. We extracted _C. albicans_ RNA in the presence or absence of PMT12-BF4
at 1 μg/mL for RNA sequencing experiments. RNAs extracted from cultures in fresh YPD medium were set as a control group for further fold-change analysis. Results of RNA sequencing revealed
that transcriptome expression of cultures treated with PMT12-BF4 showed 42 differentially expressed genes (DEGs; _P_ < 0.05). In the presence of PMT12-BF4, 34 genes were up-regulated with
a log2 fold-change ranging from 2 to 9.76, and 22 of the genes have been characterized. On the other hand, 8 genes were down-regulated with a log2 fold-change ranging from 2 to 3.73, and
all of them have been characterized. Relative expression of 3 up- and 3 down-regulated genes was confirmed by qRT-PCR (Fig. 5). Gene ontology of characterized genes found that up-regulated
genes were mainly involved in oxidizing metal ions, ferric-chelate reductase activity, iron ion transmembrane transporter, and oxidoreductase activity, while down-regulated genes were mainly
involved in oxidoreductase activity, vitamin binding and coenzyme binding (Table 3). To identify potential target(s) affected by PMT12-BF4 in _C. albicans_, we screened a deletion mutant
library comprising of 666 homozygous mutants13. Three hundred and nine out of 666 mutants showed resistance (MIC >4 μg/mL) to PMT12-BF4, and among them the functions of 139 genes were
characterized and described in the _Candida_ Genome Database (CGD) website (http://www.candidagenome.org/). Among 139 genes, 29 (20.9%) genes were involved in iron-mediated regulation or
iron-related functions, 34 (24.5%) genes were involved in hyphal growth, biofilm formation, or cell wall-related functions and the remaining genes were responsible for other functions
(Supplementary Table 1). In a comparison of RNA sequencing and mutant library screening results, 6 genes (_CFL2_, _FET3_, _XOG1_, _IFD6_, _RBT_4 and _BRG1_) were up-regulated in the presence
of PMT12-BF4, and their corresponding mutants were found to be resistant to PMT12-BF4 (Table 3 and Supplementary Table 1). Among these genes, 5 of 6 genes (_CFL2_, _FET3_, _XOG1_, _RBT4_,
_BRG1_) were iron-related, while _IFD6_ was associated with biofilm formation. IRON IONS ABOLISHED THE ANTIFUNGAL ACTIVITY OF PMT12-BF4 Our experimental results from RNA sequencing and
mutant library screening revealed that iron ions may play a role in PMT12-BF4 antifungal activity, and thus a spotting assay was performed to investigate the impact of iron ions on
antifungal activity of PMT12-BF4. Three _C. albicans_ including drug-susceptible and -resistant isolates grew normally on a YNB agar plate, but growth significantly decreased after PMT12-BF4
was added (Fig. 6). Surprisingly, _C. albicans_ isolates could be recovered from this condition after the addition of Fe2+, an absorbable form of iron ions for _C. albicans_. Meanwhile,
similar results were seen when compared to the YNB agar plates containing ciclopirox olamine, an iron ion chelator, indicating that PMT12-BF4 may function in a similar manner to the iron ion
chelator and interrupt the absorption of iron ions in _C. albicans_ (Fig. 6). PMT12-BF4 EXHIBITED MODERATE TOXICITY TO HUMAN CELL LINES To determine the cytotoxicity of PMT12-BF4 against
the human neuroblastoma cell line SK-N-SH and human embryonic kidney cell line HEK293, MTT reduction assays were conducted, and cell viability was determined after PMT12-BF4 treatment. Cell
viability of both cell lines decreased as the concentration of PMT12-BF4 increased. The viability of SK-N-SH cells was lower than 50% at 5 μg/mL (45.85%) PMT12-BF4, while that of HEK293 was
lower than 50% at 10 μg/mL (32.38%) PMT12-BF4. According to the equation obtained from linear regression analysis, the IC50 of PMT12-BF4 against SK-N-SH cells was 6.78 μg/mL, and that
against HEK293 was 10.05 μg/mL (Fig. 7). DISCUSSION A previous study showed that gemini surfactants inhibited bacterial pathogens especially Gram positive strains through their surfactant
activity and the specific cell wall construction of the pathogens14. However, few studies have discussed the activity of gemini QACs against fungal pathogens, and so far no clear mode of
action has been proposed. In this study, we found that newly synthesized PMT12-BF4 had broad-spectrum fungicidal activity, especially combating drug-resistant _C. albicans_, suggesting
PMT12-BF4 uses a mode of action distinct from current antifungal drugs. Although PMT12-BF4 exhibited antifungal activity to most fungal pathogens tested, _M. furfur_ was resistant to
PMT12-BF4, indicating that it is not a general biocidal compound and _M. furfur_ might use specific detoxification system(s) to reduce the damage caused by PMT12-BF4. The finding that the
length of the hydrocarbon chain is associated with the strength of antimicrobial activity is of interest. Bao _et al_.15 demonstrated that the hydrocarbon lengths of the side chains in QACs
could decrease the critical microcelle concentration (CMC), but the antimicrobial activity against multiple pathogenic bacteria was similar among various QACs. Thus, longer or shorter
hydrocarbon chains of QACs are not beneficial to inhibition of the microbes15. Another study showed that a histidine-based surfactant could inhibit several Gram-positive and -negative
bacteria as well as _C. albicans_, and their antimicrobial activity changed as the alkyl chain length changed. The most active compound was found in DMHNHC14, a C14 homologue. This
surfactant possessed selective activity towards bacterial membranes, and had low toxicity to erythrocytes16. Our data showed that QAC compounds possess broad-spectrum antifungal activities
against pathogenic yeasts and filamentous fungi, and demonstrated best efficacy when there were 12 hydrocarbons in both side chains. The reasons why compounds with 16 hydrocarbon chains are
not effective against _C. albicans_ and _C. tropicalis_, but effective against _C, glabrata_ and _C. neoformans_ remain unclear. A previous report showed that several promising antifungal
targets against _C. albicans_ were based on ion homeostasis, such as Cfl1 and Fet317. Our RNA sequencing results showed that several genes including iron- or copper-related functions and
heme-binding genes (_e.g_., _RBT_5, _PGA7_, _CFL_2/4/5 _etc_.) were up-regulated in _C. albicans_ under treatment with PMT12-BF4. In addition, several genes in the _FET_ and _CFL_ gene
families such as _CFL1_/_2_/_4_/_5_ and _FET3_/34 were up-regulated, indicating that PMT12-BF4 may directly or indirectly regulate these gene families. Similar results were also found in _C.
albicans_ deletion mutant library screening, such that Δ_cfl2_ and Δ_fet3_ mutants showed resistance to PMT12-BF4, indicating that the mechanisms PMT12-BF4 used to target _C. albicans_
might be associated with metal (_i.e_., iron and copper) ion homeostasis. Previous reports showed that the loss of iron uptake genes such as _FET34_, a multicopper ferroxidase induced by low
iron, could result in a filamentous growth defect in _C. albicans_18,19. In addition, deletion of _CFL1_, which encodes a protein similar to ferric reductase, decreased cell wall integrity
and filamentous growth in _C. albicans_20,21. As discussed by Puri _et al_. (2019), iron-related regulation in _C. albicans_ is mainly mediated by four regulators, Tup1, Hap43, Sfu1 and
Sef122. Our mutant library screening data demonstrated that PMT12-BF4-associated iron-related genes were regulated by these regulators, and most of these genes were repressed by Hap43,
indicating the antifungal activity of PMT12-BF4 may not directly alter the iron concentration in the environment, but instead, it possibly interferes with functions of iron regulation. On
the other hand, genes associated with hyphal development, biofilm formation and cell wall-related functions were also found from mutant library screening. We noted that an alkaline
response-transcription factor mutant _rim101_ showed resistance to PMT12-BF4, and several genes associated with cell wall integrity were regulated by Rim101, indicating that Rim101might play
a role in mediating the mode of action of PMT12-BF4 against _C. albicans_. Iron chelators could be used as antifungal agents based on their ability to disrupt iron ion homeostasis and
interfere with growth and morphogenesis in _C. albicans_23,24,25. According to our RNA sequencing data, some genes regulated by PMT12-BF4 can be also upregulated by ciclopirox olamine, an
iron chelator. Meanwhile, PMT12-BF4 also showed decreased antifungal activity against _C. albicans_ after addition of iron ions, indicating the possibility that the iron uptake activity
might be changed when PMT12-BF4 targets the pathogen. Taken together, the mode of action of PMT12-BF4 against _C. albicans_ might involve interrupting cell growth, hyphal development, and
biofilm formation, as well as interfering with iron ion homeostasis. In summary, to the best of our knowledge, this is the first report showing that a gemini QAC (_i.e_., PMT12-BF4) can
inhibit _C. albicans_ via regulating iron ion homeostasis, therefore indicating that it might be a novel antifungal agent that could be developed in the future. MATERIALS AND METHODS STRAINS
AND MEDIA The fungal pathogens used in this study are shown in Table 1. The media used in this study were YPD (1% yeast extract [Bioshop, Canada], 2% peptone [Bioshop], 2% dextrose
[Bioshop]), PDB (24 g potato dextrose broth [Himedia, India] in 1 L distilled water), RPMI 1640 medium (10.4 g RPMI 1640 powder [Sigma-Aldrich, USA], 34.5 g MOPS [3-(N-morpholino)
propanesulfonic acid, Sigma-Aldrich], 2 g dextrose, in 1 L distilled water, with pH adjusted to 7.0 with NaOH), spider medium (10 g nutrient broth [Himedia], 10 g mannitol [Panreac, Spain],
2 g K2HPO4, in 1 L distilled water, adjusted to pH 7.2 with H3PO4), YNB (0.17% yeast nitrogen base w/o amino acids [Bioshop], 0.5% (NH4)2SO4, 2% dextrose) and modified Dixon medium (36 g
malt extract [Merck, Germany], 20 g desiccated oxbile [Sigma-Aldrich], 10 mL Tween 40 [Sigma-Aldrich], 6 g peptone [Bioshop], 2 mL glycerol [Scharlab, Spain], 2 mL oleic acid
[Sigma-Aldrich], in 1 L distilled water). All media were solidified by adding 2% agar (Bioshop) if needed, except mDixon medium (1.5% agar). For synthesis reaction, N, N, N′, N′,
N′′-pentamethyldiethylenetriamine [Sigma-Aldrich], alkyl bromides [Sigma-Aldrich], 1, 4-butanesultone [Sigma-Aldrich], and acetonitrile [POCh S. A., Poland] were purchased. All compounds
were analytical reagent quality and used without further purification. SYNTHESIS PROCEDURES The synthesis reaction of PMTX-BF4 (X = 12, 16) was carried out in two steps. In the first step, a
quaternary amine with a gemini structure (PMTX) was obtained. For this purpose, a reaction was carried out according to the methods described in the literature26. N, N, N′, N′, N′′-
pentamethyldiethylenetriamine (2 g, 0.01 mol) with the appropriate alkyl bromide (0.02 mol) was heated under reflux for 8–48 h in acetonitrile. The reaction time was extended for bromides
with a shorter alkyl chain (Fig. 1a). After heating was complete, the product was crystallized, filtered and dried from the solution. The gemini surfactant obtained from the first step was
then reacted with tetrafluoroboric acid in a 1:1 molar ratio, proceeding with ion exchange (Fig. 1b). The obtained crystalline (PMTX-BF4) was washed with ethanol to remove unreacted
substrates. The products’ NMR, IR and elemental analysis results are given below: PMT12-BF4 _1, 5-bis(dodecyl)-1, 1, 3, 5, 5-pentamethyl-3-aza-1, 5-pentanediammonium ditetrafluoroborate_ 1H
NMR (CDCl3) σ = 0.889 (6 H, 2 × CH3), 1.265 (36 H, CH2), 1.746 (4 H, CH2), 2.634 (3 H, CH3), 3.180 (12 H, 4 × CH3), 3.336 (8 H, 4 × CH2N+), 3.734 (4 H, CH2N) 13C NMR (CDCl3) σ = 14.242 (2 ×
CH3), 22.689 (2 × CH2), 26.189 (2 × CH2), 29.228 (2 × CH2), 29.371 (2 × CH2), 29.682 (10 × CH2), 31.922 (2 × CH2), 41.475 (CH3), 50.056 (2 × CH2N), 50.776 (4 × CH3N+), 59.363 (2 × CH2N+),
65.907 (2 × CH2N+) IR: 2917, 2850, 1467, 1030. PMT16-BF4 _1, 5-bis(hexadecyl)-1, 1, 3, 5, 5-pentamethyl-3-aza-1,5-pentanediammonium ditetrafluoroborate_ 1H NMR (CDCl3) σ = 0.911 (6 H, 2 ×
CH3), 1.288 (52 H, CH2), 1.766 (4 H, CH2), 2.715 (3 H, CH3), 3.195 (12 H 4 × CH3), 3.390 (8 H, 4 × CH2N+), 3.784 (4 H, CH2N) 13C NMR (CDCl3) σ = 14.266 (2 × CH3), 22.709 (2 × CH2), 26.194 (2
× CH2), 29.252 (2 × CH2), 29.375 (2 × CH2), 29.778 (18 × CH2), 31.942 (2 × CH2), 41.474 (CH3), 49.991 (2 × CH2N), 50.775 (4 × CH3N+), 59.277 (2 × CH2N+), 66.164 (2 × CH2N+) IR: 2915, 2849,
1469, 1037. DETERMINATION OF THE ANTIFUNGAL ACTIVITY OF THE COMPOUND Two compounds, PMT12-BF4 and PMT16-BF4 were tested in this study. The stock solutions were prepared by dissolving each
compound powder in distilled water or dimethyl sulfoxide (DMSO, [Scharlab, Spain]) at a concentration of 5 mg/mL, and then they were kept at room temperature for further use. To determine
the antimicrobial activity, we followed the Clinical and Laboratory Standards Institute (CLSI) guidelines M27-A3 for yeasts and M38-A2 for filamentous fungi. In brief, 100 μL of serially
diluted compounds (2-fold the final concentration) were added into 100 μL cells or conidia suspensions in a 96-well polystyrene plate (Nest Biotechnology, China). The final cell
concentrations were 1.25 × 103 CFU/mL for yeasts, or 2.5 × 104 conidia/mL for filamentous fungi, while the final concentrations of tested compounds ranged from 0.125 to 64 μg/mL. The plates
were incubated for 48 h at 35 °C without shaking. When conducting this assay for _Malassezia furfur_, a yeast pathogen isolated from human dandruff, the protocol was modified slightly;
mDixon medium was used and the plate was incubated at 35 °C for 7 days27. The minimal inhibitory concentration (MIC) was defined as the lowest concentration showing no visible growth. For
fluconazole and micafungin against _C. albicans_, MIC was defined as the lowest concentration for which a prominent decrease in turbidity is observed (approximately 50% decrease as
determined visually or spectrophotometrically) as described in the CLSI protocol. Minimal fungicidal concentration (MFC) was determined after the MIC test. For each strain, 3 μL of wells
containing compounds at concentrations from 0.5 MIC (positive control) to the highest concentration (64 μg/mL) were pipetted up and down thoroughly and subcultured onto drug-free YPD agar
plates. The plates were incubated at an optimal temperature for growth of each fungal pathogen for 48 h and the MFC was determined as the drug concentration at which no colonies formed.
TIME-KILLING KINETIC ASSAY Overnight cultures of _C. albicans_ SC5314 were washed twice with ddH2O and inoculated into YPD broth to a volume of 3 mL (105 CFU/mL) with or without 4 μg/mL
PMT12-BF4. The cultures were incubated at 30 °C. At the indicated time points (0, 4, 8, 12, 24 and 48 h), a 100 μL aliquot was removed from each culture and appropriately diluted with ddH2O.
A 100 μL diluted aliquot was then plated on a fresh YPD agar plate and incubated at 37 °C for 24 h before colony count. GROWTH KINETICS ASSAY _C. albicans_ cells were grown overnight in YPD
at 30 °C, and diluted to a concentration of 1.25 × 103 CFU/mL in 200 μL YPD with 2-fold serial diluted compounds ranging from 0.125 to 64 μg/mL in a 96-well plate. Medium without compounds
was used as a positive control, while that without inoculum served as a negative control. The plate was incubated at 30 °C, and OD600 was measured at the following time points (0, 2, 4, 6,
8, 10, 12, 24 and 48 h) after incubation. Every well was mixed thoroughly with a pipette before spectrometric measurement. The average optical density (OD) value of the negative control
wells was subtracted from that of each experimental and positive control well. To calculate the relative growth, wells without compounds at 48 h were set as 100% growth compared to the
initial OD value at 0 h. GERM TUBE INDUCTION ASSAY _C. albicans_ SC5314 cells were cultured in YPD broth overnight at 30 °C, washed twice with ddH2O and diluted with 1 mL RPMI 1640 medium to
OD600 0.25 in a 12-well plate (Nest Biotechnology, China). PMT12-BF4 was then added into wells at the concentrations of 0, 0.25, 0.5 or 1 μg/mL with three replicates. The plate was
incubated at 37 °C for 3 h for germ tube induction, and observed with an inverted microscope (Olympus, Japan) at 400X magnification. Cells were photographed by a camera connected with
Olympus cellSens Entry 2.1 software. BIOFILM FORMATION ASSAY Biofilm formation assay was conducted with slight modification as described previously28,29. In brief, _C. albicans_ cells were
grown overnight in YPD at 30 °C, diluted to a 0.5 OD600 in 2 mL Spider medium in a 12-well plate (Falcon, flat bottom and non-cell tissue treated). The plates were incubated at 37 °C for 2 h
at 150 rpm agitation for initial adhesion of cells. The plates were washed with 2 mL PBS, and 2 mL of Spider medium was added with PMT12-BF4 at the indicated concentrations (0, 1, 2 or 4
μg/mL). The plates were incubated at 37 °C for 24 h at 150 rpm agitation to allow biofilm formation. After incubation, the plates were washed twice with 2 mL PBS, and stained with 2 mL 0.4%
crystal violet for 45 min. After washing with ddH2O, wells were destained with 2 mL 95% EtOH for 45 min. The content (200 μL) of each well was transferred to a new 96-well plate with
appropriate dilutions, and the optical density was measured at a wavelength of 595 nm. The mean OD595 value of negative control wells (without inoculum) was subtracted from that of each
experimental and positive control well (without compound), and all values were then normalized to the mean value of positive control wells. The biofilm formation of wells without PMT12-BF4
was set as 100%. Statistical analyses (Student’s unpaired two-tailed _t_ test) were performed with GraphPad Prism 6.0 software. Significance was set as a P value less than 0.05. SCANNING
ELECTRON MICROSCOPY Cultures of _C. albicans_ grown overnight at 30 °C in YPD medium were harvested and washed twice with PBS. The cells were resuspended in 3 mL of RPMI 1640 medium with
PMT12-BF4 at the indicated concentrations (0, 0.5, 1, or 2 μg/mL), adjusted to 105 CFU/mL, and incubated at 37 °C for 3 h. After the incubation, cells were washed twice with PBS and fixed
overnight in 2.5% glutaraldehyde in 0.1 M phosphate buffer. The samples were washed three times with 0.1 M phosphate buffer, each for 10 min, and post-fixed in 1% osmium tetroxide (OsO4) for
1 h. The post-fixed cells were washed 3 times with 0.1 M phosphate buffer to remove OsO4, and then dehydrated in ethanol in a 30% to 100% gradient (once at 30%, 50%, 70%, 85%, 90%, and 95%,
each for 10 min; twice at 100% for 20 min) and 100% acetone for 10 min. The samples were thoroughly dried in a critical point dryer with liquid CO2 (Hitachi HCP-2, Japan) and coated with
gold using an ion coater (Eiko Engineering, Japan). After processing, samples were observed and photographed in a scanning electron microscope (FEI Inspect S, USA). RNA SEQUENCING
EXPERIMENTS _C. albicans_ SC5314 cells were grown overnight in liquid YPD at 30 °C, washed twice with ddH2O, and adjusted to OD600 0.25 with 5 mL fresh YPD in the presence or absence of 1
μg/mL PMT12-BF4. The cultures were incubated at 30 °C for 3 h with shaking at 200 rpm. Cells were centrifuged at 4 °C for 10 min at 3,250 rpm, and washed with ice-cold ddH2O to discard the
medium. Total RNA was extracted using TRIzol reagent (Invitrogen, USA). Collected cells were frozen in liquid N2, and vortexed with beads. After adding 1 mL TRIzol, cultures were centrifuged
at 4 °C/12,000 _g_ for 10 min, and the supernatant was transferred to a new tube. After incubation for 5 min at room temperature, 200 μL of chloroform was added and mixed thoroughly, and
tubes were incubated at room temperature for 3 min, centrifuged at 4 °C/12,000 _g_ for 15 min, and the supernatant was transferred to a new tube. Isopropanol (500 μL) was added to the tubes,
incubated at room temperature for 10 min, centrifuged at 4 °C/12,000 g for 10 min, and the supernatant was discarded. The pellet containing RNA was washed twice with 75% ethanol and
resuspended with RNase free water. The Next Generation Sequencing (NGS) library construction using RNA was as described in a previous study30. The mRNA was enriched with oligo(dT) magnetic
beads and shortened into approximately 200-base fragments in fragmentation buffer. The first strand of cDNA was synthesized by the use of a random hexamer, buffer, dNTPs, and RNase H, and
the second strand by the use of DNA polymerase I. The double strand cDNAs were purified with magnetic beads. After end preparation and 3′ end single nucleotide adenine addition were
performed, sequencing adaptors were ligated to the fragments, and amplified by PCR. An Agilent 2100 bioanalyzer and ABI StepOnePlus real-time PCR system were used to qualify and quantify the
sample library, and the library products were sequenced via an Illumina HiSeq. 2000 instrument. REAL-TIME QRT-PCR Real-time qRT-PCR was conducted as in a previous study30. Total RNA
extracted as described above was treated with a Turbo DNA-free kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol to eliminate genomic DNA contamination, and 2 μg of
DNA-free total RNAs were reverse transcribed to cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems). A 10-μL reaction volume of the real-time PCR mixtures included
1 μL of cDNA (5 ng), 5 μL of 2 × Fast SYBR green master mix (Applied Biosystems), 0.5 μL of 10 μM forward primer, and 0.5 μL of 10 μM reverse primer. Primer pairs used in real-time PCR are
listed in Supplementary Table 2. Quantitative PCR conditions were set as follows: 95 °C for 10 min for denaturation, 95 °C for 15 s and 60 °C for 60 s (40 cycles), 95 °C for 15 s,95 °C for
60 s, and 95 °C for 15 s (melting curve). Cycle threshold (_C__T_) values were determined by a StepOnePlus system and StepOne software (v2.3), and the relative gene expressions were
calculated based on _ACT1_-calibrated and 2−ΔΔ_CT_ values. The relative expression levels of _C. albicans_ genes in the presence of 1 μg/mL PMT12-BF4 were normalized to those in the absence
PMT12-BF4, and the bar graphs were obtained using GraphPad Prism 6.0 software. Significant differences were analyzed using unpaired _t_ test (_P_ < 0.05). MUTANT LIBRARY SCREENING The
homozygous knockout mutant set of _C. albicans_ was purchased from the Fungal Genetics Stock Center (http://www.fgsc.net/candida/FGSCcandidaresources.htm), and used to screen potential
target(s) of PMT12-BF413. The set contained 666 homozygous mutants and two _C. albicans_ SN152 wild-type strains derived from strain SC5314 with auxotroph of histidine, leucine, and
arginine. Among these mutants, 316 (approximately 47%) genes were characterized. For library screening, 100 μL of YPD with 4 μg/mL PMT12-BF4 was added into a 96-well plate, and the inoculum
was transferred from stock plates of knockout mutant sets with a sterile replica plater. The inoculated plates were incubated at 30 °C for 24–48 h without shaking and mutants showing visible
growth in wells were defined as resistant strains against PMT12-BF4. DETERMINATION OF CELL TOXICITY OF PMT12-BF4 The human neuroblastoma cell line SK-N-SH was cultured in Dulbecco’s
modified Eagle’s medium (DMEM, Hyclone, USA), and the human embryonic kidney cell line HEK293 was cultured in minimum essential medium (MEM, Gibco, USA). Both media contained 10% fetal
bovine serum (Corning, USA), 2 mM L-glutamine (Hyclone, USA), and antibiotic solution of 100 U/mL penicillin G, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B. Both SK-N-SH and HEK293
cells were incubated at 37 °C under 5% CO2 and passaged every 3–4 days. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide [MTT (Thiazolyl Blue
Tetrazolium Bromide, Sigma, USA)] cell viability assay in triplicate. SK-N-SH cells were seeded at a density of 2.5 × 104 cells/mL in a 96-well plate (200 μL/well), and HEK293 cells were
seeded at a density of 105 cells/mL in a 24-well plate (500 μL/well) for 24 h before PMT12-BF4 treatment at 37 °C under 5% CO2. Subsequently, SK-N-SH cells were treated with 0, 1, 2, 3, 4,
5, 6, 8, 12, and 16 μg/mL PMT12-BF4, while HEK293 cells were treated with 0, 1.25, 2.5, 5, 10, and 20 μg/mL PMT12-BF4, and plates were incubated at 37 °C under 5% CO2 for 24 h. After the
treatment, cells were incubated with MTT solution at a concentration of 5 mg/mL for 2 h at 37 °C under 5% CO2. After removal of the medium, 200 μL of DMSO was added for optical density (OD)
measurement at 575 nm using a spectrophotometer (SpectraMax 190 UV-Vis Microplate, Molecular Devices, USA). The percentages of viable cells were calculated as [(sample OD575 nm) − (blank
OD575 nm)/(control OD575 nm) − (blank OD575 nm)] × 100(%). DATA AVAILABILITY We deposited the RNA sequences in the NCBI Gene Expression Omnibus (GSE) database under accession number
GSE129191 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129191, token: apkjqyusjhoxnwd). REFERENCES * Chang, Y. L., Yu, S. J., Heitman, J., Wellington, M. & Chen, Y. L. New
facets of antifungal therapy. _Virulence_ 8, 222–236, https://doi.org/10.1080/21505594.2016.1257457 (2017). Article CAS PubMed Google Scholar * Lee, C. Structure, conformation, and
action of neuromuscular blocking drugs. _Br. J. Anaesth._ 87, 755–769 (2001). Article CAS PubMed Google Scholar * Xiao, Y. H. _et al_. Antibacterial effects of three experimental
quaternary ammonium salt (QAS) monomers on bacteria associated with oral infections. _J. Oral. Sci._ 50, 323–327 (2008). Article CAS PubMed Google Scholar * Grant, W. M. A study of the
actions of nonaromatic quaternary ammonium compounds on the eye. _Trans. Am. Ophthalmol. Soc._ 54, 417–451 (1956). CAS PubMed PubMed Central Google Scholar * Rodrigo, G., Rodrigo, C.
& Burschtin, O. A meta-analysis of the effects of ipratropium bromide in adults with acute asthma. _Am. J. Med._ 107, 363–370 (1999). Article CAS PubMed Google Scholar * Oblak, E.
& Gamian, A. The biological activity of quaternary ammonium salts (QASs). _Postepy Hig. Med. Dosw_ 64, 201–211 (2010). Google Scholar * McDonnell, G. & Russell, A. D. Antiseptics
and disinfectants: activity, action, and resistance. _Clin. Microbiol. Rev._ 12, 147–179 (1999). Article CAS PubMed PubMed Central Google Scholar * Qibin, C. _et al_. Cationic Gemini
surfactant at the air/water interface. _J. Colloid Interface Sci._ 314, 651–658, https://doi.org/10.1016/j.jcis.2007.05.063 (2007). Article ADS CAS PubMed Google Scholar * Rosen, M. J.
_Surfactants and interfacial phenomena_. Third edn, (John Wiley & Sons, Inc., 2004). * Zana, R. Dimeric and oligomeric surfactants. _Behav. interfaces aqueous solution: a review. Adv.
Colloid Interface Sci._ 97, 205–253 (2002). CAS Google Scholar * Murguía, M. C., Machuca, L. M., Lurá, M. C., Cabrera, M. I. & Grau, R. J. Synthesis and properties of novel antifungal
gemini compounds derived from N-acetyl diethanolamines. _J. Surfactants Detergents_ 11, 223–230, https://doi.org/10.1007/s11743-008-1076-4 (2008). Article CAS Google Scholar * Zieliński,
R. _Surfaktanty. Budowa, właściwości, zastosowania_. (Wydawnictwo Uniwersytetu Ekonomicznego, 2009). * Noble, S. M., French, S., Kohn, L. A., Chen, V. & Johnson, A. D. Systematic screens
of a _Candida albicans_ homozygous deletion library decouple morphogenetic switching and pathogenicity. _Nat. Genet._ 42, 590–598, https://doi.org/10.1038/ng.605 (2010). Article CAS
PubMed PubMed Central Google Scholar * Tawfik, S. M. Synthesis, surface, biological activity and mixed micellar phase properties of some biodegradable gemini cationic surfactants
containing oxycarbonyl groups in the lipophilic part. _J. Ind. Eng. Chem._ 28, 171–183, https://doi.org/10.1016/j.jiec.2015.02.011 (2015). Article CAS Google Scholar * Bao, Y., Guo, J.,
Ma, J., Li, M. & Li, X. Physicochemical and antimicrobial activities of cationic gemini surfactants with polyether siloxane linked group. _J. Mol. Liq._ 242, 8–15,
https://doi.org/10.1016/j.molliq.2017.06.049 (2017). Article CAS Google Scholar * Bustelo, M. _et al_. Monocatenary histidine-based surfactants: Role of the alkyl chain length in
antimicrobial activity and their selectivity over red blood cells. _Colloids Surf. A: Physicochemical Eng. Asp._ 532, 501–509, https://doi.org/10.1016/j.colsurfa.2017.04.017 (2017). Article
CAS Google Scholar * Li, Y. _et al_. Promising antifungal targets against _Candida albicans_ based on ion homeostasis. _Front. Cell. Infect. microbiology_ 8, 286–286,
https://doi.org/10.3389/fcimb.2018.00286 (2018). Article CAS Google Scholar * Cheng, X. _et al_. Novel insight into the expression and function of the multicopper oxidases in _Candida
albicans_. _Microbiology_ 159, 1044–1055, https://doi.org/10.1099/mic.0.065268-0 (2013). Article CAS PubMed Google Scholar * Mamouei, Z., Zeng, G., Wang, Y. M. & Wang, Y. _Candida
albicans_ possess a highly versatile and dynamic high-affinity iron transport system important for its commensal-pathogenic lifestyle. _Mol. Microbiology_ 106, 986–998,
https://doi.org/10.1111/mmi.13864 (2017). Article CAS Google Scholar * Xu, N. _et al_. Novel role of the _Candida albicans_ ferric reductase gene _CFL1_ in iron acquisition, oxidative
stress tolerance, morphogenesis and virulence. _Res. Microbiology_ 165, 252–261, https://doi.org/10.1016/j.resmic.2014.03.001 (2014). Article CAS Google Scholar * Yu, Q. _et al_. A novel
role of the ferric reductase Cfl1 in cell wall integrity, mitochondrial function, and invasion to host cells in _Candida albicans_. _FEMS Yeast Res._ 14, 1037–1047,
https://doi.org/10.1111/1567-1364.12194 (2014). Article CAS PubMed Google Scholar * Puri, S., Kumar, R., Rojas, I. G., Salvatori, O. & Edgerton, M. Iron chelator deferasirox reduces
_Candida albicans_ invasion of oral epithelial cells and infection levels in murine oropharyngeal candidiasis. _Antimicrobial Agents Chemotherapy_ 63, e02152–02118,
https://doi.org/10.1128/AAC.02152-18 (2019). Article CAS PubMed Google Scholar * Tripathi, S. K. _et al_. Two plant-derived aporphinoid alkaloids exert their antifungal activity by
disrupting mitochondrial iron-sulfur cluster biosynthesis. _J. Biol. Chem._ 292, 16578–16593, https://doi.org/10.1074/jbc.M117.781773 (2017). Article CAS PubMed PubMed Central Google
Scholar * Prasad, T., Chandra, A., Mukhopadhyay, C. K. & Prasad, R. Unexpected link between iron and drug resistance of _Candida_ spp.: iron depletion enhances membrane fluidity and
drug diffusion, leading to drug-susceptible cells. _Antimicrobial agents chemotherapy_ 50, 3597–3606, https://doi.org/10.1128/AAC.00653-06 (2006). Article CAS PubMed Google Scholar *
Niewerth, M. _et al_. Ciclopirox olamine treatment affects the expression pattern of _Candida albicans_ genes encoding virulence factors, iron metabolism proteins, and drug resistance
factors. _Antimicrobial agents chemotherapy_ 47, 1805–1817, https://doi.org/10.1128/AAC.47.6.1805-1817.2003 (2003). Article CAS PubMed Google Scholar * Wettig, S. D., Wang, C., Verrall,
R. E. & Foldvari, M. Thermodynamic and aggregation properties of aza- and imino-substituted gemini surfactants designed for gene delivery. _Phys. Chem. Chem. Phys._ 9, 871–877,
https://doi.org/10.1039/B613269C (2007). Article CAS PubMed Google Scholar * Leong, C., Buttafuoco, A., Glatz, M. & Bosshard, P. P. Antifungal susceptibility testing of _Malassezia_
spp. with an optimized colorimetric broth microdilution method. _J. Clin. Microbiol._ 55, 1883–1893, https://doi.org/10.1128/JCM.00338-17 (2017). Article CAS PubMed PubMed Central Google
Scholar * Nobile, C. J. _et al_. A recently evolved transcriptional network controls biofilm development in _Candida albicans_. _Cell_ 148, 126–138,
https://doi.org/10.1016/j.cell.2011.10.048 (2012). Article CAS PubMed PubMed Central Google Scholar * Lohse, M. B. _et al_. Assessment and optimizations of _Candida albicans in vitro_
biofilm assays. _Antimicrob Agents Chemother_ 61, https://doi.org/10.1128/AAC.02749-16 (2017). * Yu, S. J., Chang, Y. L. & Chen, Y. L. Deletion of _ADA2_ increases antifungal drug
susceptibility and virulence in _Candida glabrata_. _Antimicrob Agents Chemother_ 62, https://doi.org/10.1128/AAC.01924-17 (2018). * Odds, F. C., Brown, A. J. & Gow, N. A. _Candida
albicans_ genome sequence: a platform for genomics in the absence of genetics. _Genome Biol._ 5, 230, https://doi.org/10.1186/gb-2004-5-7-230 (2004). Article PubMed PubMed Central Google
Scholar * White, T. C. Increased mRNA levels of _ERG16_, _CDR_, and _MDR1_ correlate with increases in azole resistance in _Candida albicans_ isolates from a patient infected with human
immunodeficiency virus. _Antimicrob. Agents Chemother._ 41, 1482–1487 (1997). Article CAS PubMed PubMed Central Google Scholar * Garcia-Effron, G., Park, S. & Perlin, D. S.
Correlating echinocandin MIC and kinetic inhibition of _fks1_ mutant glucan synthases for _Candida albicans_: implications for interpretive breakpoints. _Antimicrob. Agents Chemother._ 53,
112–122, https://doi.org/10.1128/AAC.01162-08 (2009). Article CAS PubMed Google Scholar * Butler, G. _et al_. Evolution of pathogenicity and sexual reproduction in eight _Candida_
genomes. _Nat._ 459, 657–662, https://doi.org/10.1038/nature08064 (2009). Article ADS CAS Google Scholar * Dujon, B. _et al_. Genome evolution in yeasts. _Nat._ 430, 35–44,
https://doi.org/10.1038/nature02579 (2004). Article ADS Google Scholar * Perfect, J. R., Ketabchi, N., Cox, G. M., Ingram, C. W. & Beiser, C. L. Karyotyping of _Cryptococcus
neoformans_ as an epidemiologic tool. _J. Clin. Microbiol._ 31, 3305–3309 (1993). Article CAS PubMed PubMed Central Google Scholar * Castella, G., Coutinho, S. D. & Cabanes, F. J.
Phylogenetic relationships of _Malassezia_ species based on multilocus sequence analysis. _Med. Mycol._ 52, 99–105, https://doi.org/10.3109/13693786.2013.815372 (2014). Article CAS PubMed
Google Scholar * Nierman, W. C. _et al_. Genomic sequence of the pathogenic and allergenic filamentous fungus _Aspergillus fumigatus_. _Nat._ 438, 1151–1156,
https://doi.org/10.1038/nature04332 (2005). Article ADS CAS Google Scholar * O’Donnell, K. _et al_. Genetic diversity of human pathogenic members of the _Fusarium oxysporum_ complex
inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and
nosocomial origin. _J. Clin. Microbiol._ 42, 5109–5120, https://doi.org/10.1128/JCM.42.11.5109-5120.2004 (2004). Article CAS PubMed PubMed Central Google Scholar * Ma, L. J. _et al_.
Comparative genomics reveals mobile pathogenicity chromosomes in _Fusarium_. _Nat._ 464, 367–373, https://doi.org/10.1038/nature08850 (2010). Article ADS CAS Google Scholar Download
references ACKNOWLEDGEMENTS We are grateful to Miranda Loney for language editing and Technology Commons, College of Life Science, National Taiwan University (Taiwan) for SEM observation.
This work was financially supported by grants 106-2923-B-002-001-MY3, 107-2923-B-002-001-MY3 from the Ministry of Science & Technology in Taiwan and grant PL-TW IV/2/2017 from the
National Centre of Research and Development in Poland. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Plant Pathology and Microbiology, National Taiwan University, 10617,
Taipei, Taiwan Li-Hang Hsu, Shih-Cheng Wang, Tang-Long Shen & Ying-Lien Chen * Department of Technology and Instrumental Analysis, Poznan University of Economics and Business, Poznan,
Poland Dobrawa Kwaśniewska & Daria Wieczorek Authors * Li-Hang Hsu View author publications You can also search for this author inPubMed Google Scholar * Dobrawa Kwaśniewska View author
publications You can also search for this author inPubMed Google Scholar * Shih-Cheng Wang View author publications You can also search for this author inPubMed Google Scholar * Tang-Long
Shen View author publications You can also search for this author inPubMed Google Scholar * Daria Wieczorek View author publications You can also search for this author inPubMed Google
Scholar * Ying-Lien Chen View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS L.H.H. designed and performed experiments; analyzed and
interpreted data and wrote the manuscript. D.K. synthesized Gemini quaternary ammonium compounds and wrote the manuscript. SCW performed experiments related to mammalian cell toxicity and
wrote the manuscript. T.L.S. supervised and designed the experiments of mammalian cell toxicity and wrote the manuscript. D.W. synthesized Gemini quaternary ammonium compounds and wrote the
manuscript. Y.L.C. supervised, designed experiments; analyzed and interpreted data and wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Daria Wieczorek or Ying-Lien Chen. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in
published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hsu, LH., Kwaśniewska, D., Wang, SC. _et al._ Gemini quaternary ammonium compound
PMT12-BF4 inhibits _Candida albicans_ via regulating iron homeostasis. _Sci Rep_ 10, 2911 (2020). https://doi.org/10.1038/s41598-020-59750-5 Download citation * Received: 03 November 2019 *
Accepted: 30 January 2020 * Published: 19 February 2020 * DOI: https://doi.org/10.1038/s41598-020-59750-5 SHARE THIS ARTICLE Anyone you share the following link with will be able to read
this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative