Construction of a novel signature and prediction of the immune landscape in gastric cancer based on necroptosis-related genes
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Necroptosis, a type of programmed cell death, has become a potential therapeutic target for solid tumors. Nevertheless, the potential roles of necroptosis-related genes (NRGs) in gastric
cancer (GC) remain unknown. The objective of the present study was to create a necroptosis-related prognostic signature that can provide more accurate assessment of prognosis in GC. Using
The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) data, we identified differentially expressed NRGs. Univariate analysis and Lasso regression were performed to determine the
prognostic signature. Risk scores were calculated and all GC patients were divided into high- and low-risk score group according to the median risk score value. The robustness of this
signature was externally validated with data from GSE84437 cohort (n = 431). Survival analysis revealed high-risk score patients had a worse prognosis. Results evidenced that the signature
was an independent prognosis factor for survival. Single-sample sequence set enrichment analysis (ssGSEA) exhibited different enrichment of immune cells and immune-related pathways in the
two risk groups. Furthermore, a predictive nomogram was generated and showed excellent predictive performance based on discrimination and calibration. In addition, the risk score positively
correlated with tumor mutational burden and was associated with sensitivity to multiple anti-cancer drugs. Overall, our work demonstrates a close relationship between necroptosis and the
prognosis of GC. The signature we constructed with potential clinical application value, can be used for prognosis prediction and being a potential therapeutic responses indicator in GC
patients.
Gastric cancer (GC) is a leading contributor to global cancer mortality1. Attributed to the lack of early and effective detection methods, GC with early metastasis is characterized by poor
prognosis, unsatisfactory treatment effect2,3. To date, surgical resection is currently the therapy of choice for the most of patients4. For most early stages of GC patients, clinical
symptoms are atypical and the signs are not obvious, causing delayed diagnosis, missed diagnosis and loss of chance for surgical excision. Therefore, this raises an urgent need for
developing more effective diagnostic, prognostic and therapeutic biomarkers.
As early as 2005, Alexei discovered a programmed cell death pattern that is different from normal cell apoptosis: necroptosis, and then be confirmed as a unique pattern of cell death5.
Unlike other programmed apoptosis, necroptosis is a form of regulated cell death that is characterized by formation of the necrosome complex based on swelling injury6. Necroptosis may
accelerate cancer cell death or enhance the sensitivity of tumor cells to anti-cancer treatment7,8,9,10. Research has demonstrated that necroptosis is able to overcome resistance to cancer
drugs mediated by P-glycoprotein, Bcl-2, and Bcl-xL in cancer cell lines11. Zhao et al. found that necroptosis-related lncRNAs could predict prognosis and help make a distinction between the
cold and hot tumors for improving individual therapy in GC12. All these results indicated necroptosis may be a potent therapeutic target for the treatment of cancer. Nevertheless, the
precise role of necroptosis-related genes (NRGs) in GC still not clear. Hence, understanding the impact of NRGs on GC development may provide potential prognostic biomarkers and therapeutic
targets and guide immunotherapy strategies for GC.
In this study, we aimed to develop a necroptosis-related prognostic signature (NRGsig) with guiding significance for GC prognosis and immunotherapy. We successfully divided GC patients into
two necroptosis-related molecular subtypes with diverse clinical outcomes and tumor microenvironment (TME) infiltration characteristics. Furthermore, we created the NRGsig to quantify the
level of necroptosis and assist prognosis assessment and therapeutic decision making for individual patients with GC.
By mining the Gene Set Enrichment Analysis (GSEA) (http://www.gsea-msigdb.org/gsea/index.jsp), we acquired the necroptosis gene set M24779.gmt, which contains eight necroptosis-associated
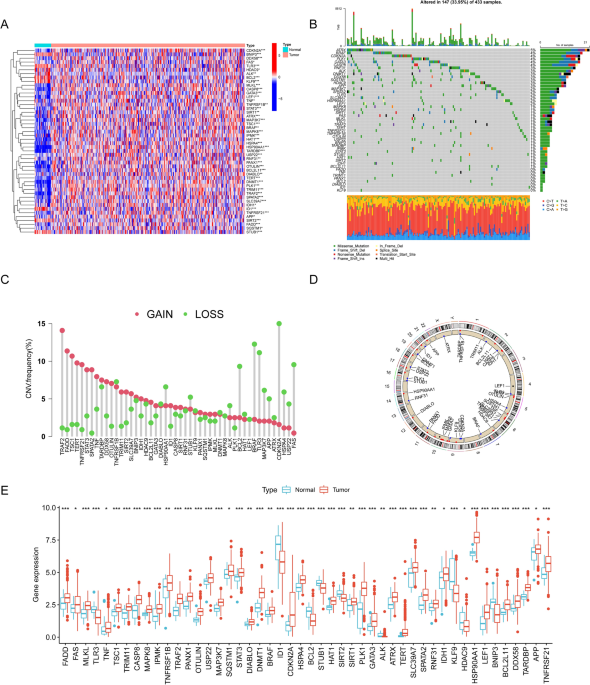
genes. After an extensive literature search about necroptosis, we ultimately identified 67 NRGs13,14,15 (details are presented in Table S1). Differentially expressed NRGs (DENRGs) in tumor
and normal tissues in the TCGA-GC cohort were screened using the “limma” package, with p