Mitogenome selection in the evolution of key ecological strategies in the ancient hexapod class collembola
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT A longstanding question in evolutionary biology is how natural selection and environmental pressures shape the mitochondrial genomic architectures of organisms. Mitochondria play a
pivotal role in cellular respiration and aerobic metabolism, making their genomes functionally highly constrained. Evaluating selective pressures on mitochondrial genes can provide
functional and ecological insights into the evolution of organisms. Collembola (springtails) are an ancient hexapod group that includes the oldest terrestrial arthropods in the fossil
record, and that are closely associated with soil environments. Of interest is the diversity of habitat stratification preferences (life forms) exhibited by different species within the
group. To understand whether signals of positive selection are linked to the evolution of life forms, we analysed 32 published Collembola mitogenomes in a phylomitogenomic framework. We
found no evidence that signatures of selection are correlated with the evolution of novel life forms, but rather that mutations have accumulated as a function of time. Our results highlight
the importance of nuclear-mitochondrial interactions in the evolution of collembolan life forms and that mitochondrial genomic data should be interpreted with caution, as complex selection
signals may complicate evolutionary inferences. SIMILAR CONTENT BEING VIEWED BY OTHERS POSITIVE SELECTION OVER THE MITOCHONDRIAL GENOME AND ITS ROLE IN THE DIVERSIFICATION OF GENTOO PENGUINS
IN RESPONSE TO ADAPTATION IN ISOLATION Article Open access 08 March 2022 AMERICAN MASTODON MITOCHONDRIAL GENOMES SUGGEST MULTIPLE DISPERSAL EVENTS IN RESPONSE TO PLEISTOCENE CLIMATE
OSCILLATIONS Article Open access 01 September 2020 MITOGENOMIC RESOLUTION OF PHYLOGENETIC CONFLICTS AND ADAPTIVE SIGNATURES IN FELIFORM CARNIVORANS Article 28 May 2025 INTRODUCTION
Understanding the interplay between genomes, morphology, functional ecology, and biogeography in driving organismal evolution has become central to answering fundamental questions in
biology, both from historical (e.g. past speciation) and contemporary (e.g. evolutionary responses to climate change) viewpoints. Making sense of the immense complexity of life has proven a
recalcitrant challenge to biologists. Additional lines of evidence pertaining to taxonomy (the fossil record in conjunction with morphological, behavioural, physiological, and molecular
data) enhance our ability to piece together the diversification of life on Earth1,2,3,4. A central question in molecular evolution revolves around how natural selection influences the
genomic architectures of organisms5,6,7,8,9,10. Environmental pressures drive functional and morphological changes, which are underpinned by genetic variation and, therefore, will manifest
as signatures of selection on the genome that can be used to retrace the evolutionary history of organisms and better predict future adaptive evolutionary trajectories. The mitochondrial
genome comprises 13 protein-coding genes (PCGs), which, together with many nuclear genes, encode the proteins that make up the subunits of the four complexes involved in the oxidative
phosphorylation chain11,12. Due to the crucial role of PCGs in the respiratory chain, these genes are functionally highly constrained5. An evaluation of signals of past selection on these
genes, therefore, provides insights into the adaptive evolution and metabolic requirements of organisms, given that they are responsible for 95% of the energy metabolism and heat production
in cells11,13,14. Mitochondrial genes are some of the most commonly used genetic markers, partly because they provide unique perspectives on population genetic variation and structure due to
maternal inheritance and a general lack of recombination15,16,17, and are frequently used to answer questions related to historical processes and geographical structure13,14,18,19. However,
these inferences often assume that the evolution of this genome is neutral20,21, an idea that has been challenged22,23,24,25,26. It is now widely accepted that the mitogenome is under
selection due to its pivotal role in respiration and energy production, and the evolutionary constraints resulting from the co-evolution of nuclear and mitochondrial genes that are involved
in these processes11,12,15,16,17,25. Additionally, positive selective sweeps, linked to thermal and aerobic respiratory adaptations, are well documented for mitochondrial genes across
various taxa11,13,14,27,28,29,30,31,32. However, the investigation of positive selection on moderately conservative genomic regions, without an assessment of the functional consequence of
amino acid mutations, is not adequate to consider the mitochondrial genome in its entirety to be under positive selection linked to adaptive processes13. Therefore, we identify both signals
of positive selection and the functional and adaptive significance of amino acid mutations on the mitochondrial genomes of ancient soil-dwelling invertebrate taxa. Given the influence of
heterogeneous soil environments on soil biota community assemblages33,34,35, and that positive selection on the mitogenome is linked to thermal and aerobic respiratory adaptations, we sought
to explore the effects of signatures of past selection on the mitogenome of one of the most ancient soil-dwelling invertebrate taxonomic groups, Collembola. Commonly referred to as
springtails, this group of primitive, wingless, and largely terrestrial hexapods is regarded as among the first aquatic arthropods to have colonised land36, with evolutionary roots that
predate the diversification of insects37,38,39,40. Indeed, the earliest known hexapod fossil (from the early Devonian period, ca. 400 Ma) is a collembolan41,42. This group's biology and
ancient evolutionary history provide unique opportunities to understand the environmental and physiological mechanisms governing the evolution of early terrestrial life. Collembolans
currently exploit a myriad of niches across all continents, including Antarctica40,43,44. They typically reside within, or are closely associated with soils, and are considered essential
components of soil ecosystems due to their influence on soil microbial communities, their role in nutrient recycling, and their importance as prey to a wide range of predatory
arthropods45,46,47,48. Of particular interest is the preference of various springtails for different habitats. Springtail species broadly fall into six different ecological life form
categories, each displaying a unique stratification preference. These are atmobiotic (large species that inhabit macrophytes (grasses, bushes, tree trunks and branches)); epiedaphic (species
found in vegetated habitats or the upper litter layer above the soil); hemiedaphic (which includes species that occupy decomposed litter or rotten wood on the soil surface); euedaphic
(‘true’ soil living species that occupy the upper mineral-rich layers of the soil); myrmecophilous (describes species found in ant nests), and hydrophilous (hereafter referred to as aquatic
for simplicity; species that live on the surfaces of, or are closely associated with, freshwater bodies)34,43,47,49,50. Strong correlations exist between the life forms of springtails and
their morphologies, suggesting that life forms are subject to selective forces exerted by their environment43,45,46,47,51,52,53. Additionally, springtails also exhibit highly specific
habitat preferences for different soil types, soil chemistries, habitat types, and stratification within a given habitat47,49,52. It is remarkable that these morphological designs have
probably persisted unchanged for millions of years in many instances. Indeed, springtail fossils dating back as far as the Eocene (ca 40–50 Ma) can be assigned to extant genera and, in some
cases, even extant species54. This extreme morphological conservatism suggests that current morphospecies inventories underestimate true collembolan diversity. Not surprisingly, genetic
evidence has shown that cryptic diversity is pervasive in this group, with current collembolan species richness thought to underestimate the actual number of species by at least an order of
magnitude40,55,56. Therefore, the low number of known springtail species is not an inherent characteristic of the group, but rather the result of under sampling, outdated taxonomy, and a
lack of taxonomic expertise56. While Collembola may be dwarfed by other hexapod groups, particularly insects, it still harbours immense diversity40. Given that Collembola constitutes
ecologically important taxa within soil environments48, which exhibit distinct microhabitat stratification that is driven by feeding strategies, food availability, and soil type and
chemistry47,49,50,52,57, we sought to identify mitochondrial genes that are under positive selection for adaptations to these microhabitats. Additionally, given that metabolic activity and
oxygen consumption differ depending on ecological life form58,59, we investigated whether signals of selection on the mitochondrial genome are associated with the evolution of life form
traits (habitat preferences), in the context of environmental change (oxygen levels/metabolic requirements and vegetation alterations) that emerged during the group's evolutionary
history. If this is the case, we expect substantial signals of selection to have accumulated along phylogenetic branches representing shifts in life forms compared to sister branches where
the original life form was maintained. RESULTS DATED PHYLOGENY AND ANCESTRAL STATE RECONSTRUCTION Our dated mitogenomic phylogeny demonstrates that Collembola first diverged sometime in the
early Devonian (ca. 410 Ma), with most diversification taking place in the Triassic to Cretaceous periods (ca. 250–60 Ma) (Fig. S1). To identify whether a link exists between signals of
mitochondrial DNA selection and collembolan life form (see Dataset S1), we assessed phylogenetic relationships between 32 collembolan and three outgroup taxa (see Table S1). We assigned a
life form trait (i.e. aquatic, epiedaphic, euedaphic, hemiedaphic, and myrmecophilous) to the ecology (i.e. habitat preference) of each springtail species included in this study (see Dataset
S1 for more detail). We reconstructed the ancestral ecological life form state for the collembolan clade using three different methods (parsimony, maximum likelihood, and Bayesian
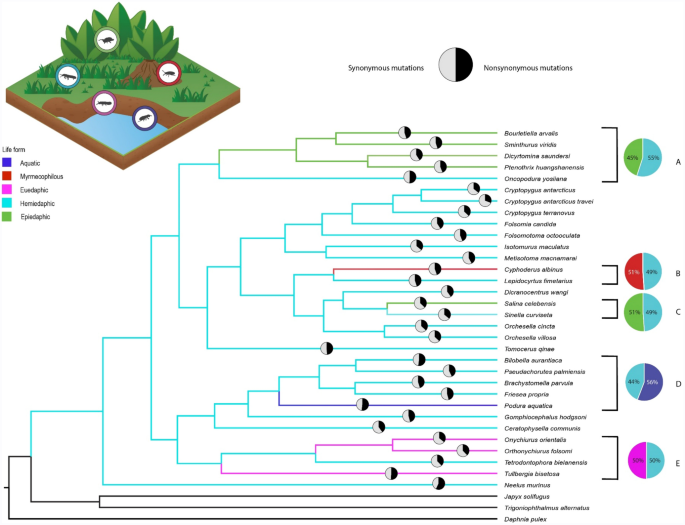
inference), all of which identified hemiedaphic as the ancestral character (see Fig. 1). Across the collembolan phylogeny, five independent life form shifts occurred from the hemiedaphic to
alternative life forms (see Fig. 1). For the terminal nodes, the most common ecological life form was hemiedaphic (22 taxa), followed by epiedaphic (five taxa), euedaphic (three taxa),
myrmecophilous (one taxon), and aquatic (one taxon). To assess phylogenetic conservatism and homoplasy across the phylogeny, we calculated the retention index (RI)60 for each life form
trait, which was 0.75 for epiedaphic, 0.36 for hemiedaphic, 0.33 for euedaphic, and 0 for both myrmecophilous and aquatic. SELECTION ANALYSIS To assess signals and direction of selection at
different loci (i.e. positive vs purifying selection), we used the codon-based maximum likelihood (CodeML) algorithm to determine the ratio of nonsynonymous to synonymous mutations (ω =
dN/dS), and the number of nonsynonymous mutations as a proportion of the total number of mutations (dN/(dN + dS))61,62,63,64. Site model selection analyses indicated overall positive
selection across the phylogeny, shown by significant likelihood ratio tests (LRT) for the comparative site models, however, with no nucleotide positions under positive selection (Bayes
Empirical Bayes score (BEB) < 0.9562) (Table S2). The comparison between the numbers of synonymous to nonsynonymous mutations across the terminal nodes of the phylogeny was significantly
different (paired _t_ test: _t_415 = − 9.48, _p_ < 0.05). The number of nonsynonymous mutations as a proportion of the total number of mutations revealed no strong evidence for a link
between life form transitions and the number of nonsynonymous mutations. Departures from the expectation that synonymous mutations are more common than nonsynonymous mutations65 were found
in several taxa (e.g. _Bilobella aurantiaca_, _Neelus murinus_, _Oncopodura yosiiana_, _Podura aquatica_, and _Tullbergia bisetosa_) (see black and white pie charts in Fig. 1 and Table S3
for proportion values > 0.50). However, only the life histories of _Podura aquatica_ and _Tullbergia bisetosa_ deviated from the hemiedaphic ancestral state. The large proportions of
nonsynonymous mutations are likely due to the older ages of these branches and illustrate that the proportion of nonsynonymous mutations increases as a function of time for the taxa with the
ancestral state compared to those that represent life form shifts (i.e. the alternative state) (ancestral state: R2 = 0.52; alternative state: R2 = 0.82; also see Fig. 2). The 95%
confidence intervals of these regressions broadly overlap, and the two models were not significantly different (_p_ > 0.05). This indicates that no significant difference was found in the
proportion of nonsynonymous mutations in relation to branch length for taxa that retained the ancestral state and for those whose ecological life form shifted. The relationships
(represented as coloured pie charts in Fig. 1) between the taxa that represent the life form transitions to alternative life forms and the respective sister taxon/taxa (ancestral state, i.e.
hemiedaphic), again revealed that the longer branches have the greatest number of nonsynonymous mutations in comparison to the sister clades (see coloured pie charts A and D). No
significant difference was found within pie charts B, C, and E, which show that the proportion of mutations for the branches that represent the alternative life form vs. ancestral state (51%
for pie charts B and C, and 50% for pie chart E; paired _t_ test: _t_4 = 0.23, _p_ > 0.05) (Fig. 1). Evidence for purifying (ω < 1) or diversifying selection (ω > 1) was determined
for each of the terminal branches, and a total of 19 taxa had mitogenomes with signatures of purifying selection, while the mitogenomes of 11 taxa showed signs of diversifying selection,
and two taxa had mitogenomes that satisfied the assumptions of neutrality (see Table S3). The proportion of changes per gene and per taxon reveal that _atp8_ exhibits the highest number of
changes for the majority of the taxa (Figs. S2, S3; Table S4). Following _atp8_, all genes from complex I (i.e. _nad1_–_nad6_) had a large proportion of nonsynonymous mutations. Of the 13
PCGs, eight genes were identified as experiencing purifying selection, while five genes were identified as experiencing diversifying selection, with _atp8_ being under particularly strong
diversifying selection (ω > 3.0) (Table S4). At a complex level, complex V (ATPase) revealed the highest percentage of nonsynonymous mutations, followed by complex I (NDs), complex IV
(COXs), and complex III (_cytb_) (Fig. S3), which follows the estimated mutation rate of these complexes (i.e. ATPase > ND > COX > CYTB)66. The average value of ω for all genes
across all terminal nodes was 0.96, which indicates that, overall, Collembola mitogenomes are experiencing slight purifying selection or no selection (i.e. selective neutrality). RADICAL
CHANGES IN PHYSICOCHEMICAL PROPERTIES OF AMINO ACIDS The impact of positive selection on the physical and chemical properties of 31 amino acids was evaluated for each of the 13 PCGs by
detecting observed changes inferred by the phylogenetic tree under neutral conditions. Both positive and negative significant z-scores on the physicochemical properties of amino acids were
found across the tree. Most physicochemical properties indicated that the radical changes were under purifying selection, with occasional radical changes under positive selection (Fig. S4).
We detected two physicochemical properties—equilibrium constant (ionisation of COOH) and solvent accessible reduction ratio—with significant radical changes (z > 3.09; _p_ < 0.001),
indicating positive/diversifying selection for all 13 PCGs, while a third physicochemical property, buriedness, is under positive/diversifying selection for 10 PCGs (except _cytb_, _nad3_,
and _nad4l_), with _atp8_ having the highest z-score. DISCUSSION Using a comprehensive mitogenome dataset for springtails (32 species from all four orders), we determined the ancestral
ecological life form state for Collembola. We then used this dataset to assess whether a correlation exists between signatures of mitogenome selection and the evolution of novel life forms.
We show that the ancestral ecological life form state of Collembola is hemiedaphic, with epiedaphic life forms first emerging in the early Permian, roughly 290 Ma. Even though several
previous studies have identified links between Collembolan mitogenomes and ecology, morphology, and physiology50,53,67,68,69,70,71, we found no strong evidence for a link between signatures
of mitochondrial genome selection and life form evolution in Collembola. Our mitogenomic phylogeny is broadly consistent with those presented in earlier studies37,38,39,72,73,74,75. The
first phylogenetic split within Collembola occurred during the early Devonian (ca. 410 Ma), with most diversification taking place in the Triassic to Cretaceous periods (ca. 250–60 Ma). It
is not surprising, therefore, that Collembola fossils preserved in amber and dating back to the Eocene (50-40 Ma) can be assigned to extant taxa with high certainty54, demonstrating the
ultraconservative nature of life forms and basic body plans of the group. Ancestral state reconstructions showed the ancestral ecological life form of collembolans to be hemiedaphic. Our
finding contrasts with the assumption of a euedaphic origin of springtails proposed by D’Haese36,76. Additionally, we found that the epiedaphic life form evolved at least twice, first during
the origin of Symphypleona (Fig. S1), approximately 290 Ma, and then again in the mid-Cretaceous with the emergence of the evolutionary lineage represented by _Salina celebensis_, roughly
115 Ma. During the Permian period, roughly 300 Ma, seed-producing gymnosperms such as gnetophytes, Pinaceae, and cupressophytes diverged from cycads and conifers77. Symphypleona show a
habitat preference for Mediterranean-type shrublands, pine plantations, mixed forests, bushes, grasslands, and leaf litter (see Supplementary Information Dataset S1). This suggests that the
emergence of major plant lineages, such as gymnosperms, created new ecological niches for the epiedaphic Symphypleona to exploit and diversify. The extant _Bourletiella arvalis_ and
_Sminthurus viridis_, however, show a habitat preference for grasslands, although these habitats would have only become available much later in the Cenozoic78,79,80. It is, therefore,
reasonable to assume that the evolutionary lineage represented by these species would have occupied gymnosperm ecological analogs until their current niches became available. Unsurprisingly,
the morphological features of these taxa are characteristic of the epiedaphic life form47,49,81. Additionally, our analyses illustrate that the euedaphic life form evolved at least twice,
first when _Tullbergia bisetosa_ evolved, roughly 287 Ma, and secondly during the origin of _Onychiurus orientalis_ and _Orthonychiurus folsomi_ (approximately 192 Ma). A shift from
hemiedaphic to the myrmecophilous and aquatic life forms was also identified in the phylogeny. The origin of _Cyphoderus albinus_ and the myrmecophilous life form occurred approximately 170
Ma, coinciding with the evolution of ants (Formicidae) around 168 Ma82,83. The aquatic life form first emerged about 225 Ma with the divergence of the lineage represented by _Podura
aquatica_. The hypothesis of a semi-aquatic or aquatic life form as the ancestral state of Collembola was formulated and tested by D’Haese36,76, who found that the evolution of terrestrial
to aquatic habitats, in a physiological context, is indeed more plausible and that an aquatic life form can be viewed as a “rare secondary acquisition”36,43,76. We deem the five life forms
we have identified to be somewhat homoplastic (see Yu et al.53) best illustrated by the epiedaphic and euedaphic forms that independently originated twice within the group. Additionally, the
myrmecophilous and aquatic forms are homoplastic or autapomorphic, as these traits are unique to single taxa. There is also a great deal of ecological conservatism for the hemiedaphic trait
where most extant taxa have retained the ancestral state. While there appeared to be a general trend for taxa on longer phylogenetic branches to have a greater overall proportion of
nonsynonymous mutations, we found no consistent relationship between life form shifts and the proportion of nonsynonymous mutations on these branches. Furthermore, even when comparing
individual genes, no correlations were found between the proportion of nonsynonymous changes and the evolution of new life forms (Fig. S2). This may seem surprising given the pivotal role
that mitochondria play in aerobic metabolism, and given that different life forms are constrained in different ways metabolically. It should be noted, however, that for a group as ancient as
Collembola, the many adaptive nuclear changes that produced the conspicuous ecological changes upon which life forms are classified may have obscured mitochondrial signatures of selection
related to the evolution of life forms. While our expectations pertaining to selection were premised on non-overlapping habitat stratification preferences, it should also be noted that
interactions between collembolans and their environments are complex, and apparently divergent life forms may impose similar functional constraints on the organism, thereby confounding
signatures of selection84. Even though mitochondrial protein coding genes are typically believed to have faster mutation rates than nuclear genes16, most of these mutations are probably
synonymous85,86, and thus functionally inconsequential (but see Lawrie et al.87). Our results support this idea, with more taxa and more genes under purifying selection than under
diversifying selection (Tables S3 and S4). However, selection on the mitochondrial genome appears to be largely neutral due to the random fixation of selectively neutral mutations21. Given
the crucial role of mitochondria in aerobic metabolism, functional requirements likely constrain the molecular evolution of mitogenomes, and this is perhaps why many studies have identified
purifying selection88,89,90. It makes sense that strong functional constraints would mean that any deleterious mutation that arises would be rapidly removed from the population via purifying
selection. Additionally, genetic drift plays a major role in structuring genetic diversity, and its effects on mitochondrial genomes are stronger than on nuclear genomes due to differences
in effective population sizes between them and the uniparental inheritance and lack of recombination within the mitochondrial genome16,17,21. When radical amino acid changes are favoured by
selection, local directional shifts in the structure and function of proteins evolve. The physicochemical properties under positive selection for all PCGs were equilibrium constant
(ionisation of COOH) and solvent accessible reduction ratio, while buriedness was under positive selection for 10 of the 13 PCGs. Positive selection for equilibrium constant is suggestive of
adaptations to increased oxygen levels and metabolic requirements66, while positive selection for solvent accessible reduction ratio may lead to bulkier proteins, which increases the space
for active site formation66. Buriedness increases protein stability, and thus optimal protein function, in the context of hydrophobicity, whereby some residues are buried on the interior of
the globular protein91,92. These results illustrate the importance of these properties in the context of environmental conditions (oxygen levels and metabolic requirements). The
mitochondrial genome has long been considered to be selectively neutral22,23,24,25, but a plethora of studies have illustrated that it may be under strong positive selection as a result of
thermal and aerobic respiratory adaptations11,13,14,27,28,29,30,31,32. Here, we investigated whether a link exists between mitochondrial genome selection and the evolution of ecological life
forms in Collembola. The expected outcome of such a link would be evidence for strong signals of selection at branches representing the evolution of new life forms, even if mitochondrial
genes themselves are not the primary drivers of ecophysiological adaptation. Our selection results reveal that, overall, positive selection is present across the collembolan phylogeny, best
illustrated by the presence of positive selection on three amino acid physicochemical properties which likely play instrumental roles in adaptive responses to environmental conditions
(oxygen levels and metabolic requirements). However, the only major trend identified here was that the proportion of nonsynonymous mutations increased with time, with longer branches (i.e.
older taxa) having a greater proportion of nonsynonymous mutations than shorter branches. In addition to investigating selection associated with life form evolution, we assessed whether
there was a link between signatures of selection across a latitudinal gradient from where each species can be found. This was done in two ways; first by selecting species across all
latitudes but where their ranges overlapped, and secondly by selecting species that had no overlapping distributions. As was the case for life form shifts, we found no clear correlation
between the number of nonsynonymous mutations and latitudinal gradients for either analysis. The lack of a correlation between life form shifts and mitogenome selection may hint at nuclear
selection, and suggests that nuclear-mitochondrial interactions are of greater importance in the evolution of life form shifts than selection pressures on the mitochondrial genome alone. Our
findings raise some precautionary flags in that selection, where present, is more complex than generally assumed, given the complex interactions of the mitochondrial and nuclear genomes.
Importantly, evidence for strong selection on the mitochondrial genome cannot simply be extrapolated to reflect adaptation to changing environmental conditions. METHODS PHYLOMITOGENOMIC
ANALYSIS Phylomitogenomic relationships between Collembola taxa based on available data on GenBank (May 2020) were reconstructed using the protein-coding genes (PCGs) of 32 partial or
complete Collembola mitogenomes, as well as three outgroup taxa (see Supplementary Information Table S1). Each PCG was aligned using the MUSCLE algorithm in MEGA X93, followed by
concatenating the alignments into a single dataset. Substitution saturation was assessed on each codon position using DAMBE v7.3.594, which revealed high substitution saturation, likely due
to highly divergent taxa95,96,97. As a result, we took a conservative approach and used the amino acid sequences of each PCG for our phylogenetic reconstructions. Each PCG was independently
translated into amino acid sequences using the invertebrate mitochondrial genetic code, and were then aligned in MEGA X using the MUSCLE algorithm, followed by manual inspection. Bayesian
phylogenetic inferences were executed in BEAST v2.6.398 by linking site and clock models as well as trees. This was done because of the largely uniparental inheritance of mitochondrial
genomes and a lack of recombination15,16,17,27. A discrete trait was added to define the ecological life form of each species (see Supplementary Information Dataset S1 for more detail).
Given the role of mitochondria in different environments, and rather than linking life forms to morphological characteristics, we linked the life forms (i.e. aquatic, myrmecophilous,
euedaphic, hemiedaphic, and epiedaphic) to the ecology (i.e. habitat preferences) of each springtail species included in this study however, with the exception of three taxa; _Onychiurus
orientalis_, _Orthonychiurus folsomi_, and T_ullbergia bisetosa_, which were assigned based on morphology. These species, although ecologically hemiedaphic47,49,50 (see Dataset S1 for
ecological life forms), differ from other hemiedaphic taxa by having all three major euedaphic morphological characteristics (no eyes, no pigment, and no furca), and are thus categorised as
“euedaphomorphic” sensu lato99. As such, these taxa were assigned to the euedaphic life form which aligns to a peculiar ecology within litter (i.e. lower dispersal ability and associated
traits) that differs from the other hemiedaphic taxa. The mitochondrial REV + Г model was selected, with an uncorrelated relaxed lognormal clock and a birth–death tree prior. Monophyly was
constrained for several clades based on a consensus tree constructed using the NCBI taxonomy database in PhyloT v2 (https://phylot.biobyte.de/), and visualised in iTOL100. These were (a) all
taxa (to calibrate the root of the tree, as described below), (b) the Hexapoda clade (all taxa excluding the distantly related _Daphnia pulex_) to calibrate the split between Collembola and
other basal hexapod taxa (_Japyx solifugus_ and _Trigoniophthalmus alternatus_) and Collembola as described below, and (c) a clade that excluded all non-Collembola taxa. Monophyly was not
constrained for each collembolan order (Poduromorpha, Entomobryomorpha, Neelipleona, and Symphypleona) since monophyly is not always supported for these groups101,102,103. Considering that
some collembolan fossils have been assigned to contemporary species despite being more than 20 Ma old, dating recent nodes using fossil data can be challenging for these taxa. We therefore
used secondary calibration points from Leo et al.104 and included calibration points that matched the two deepest fossil-calibrated nodes. To ensure that our calibration points matched the
95% highest probability density (HPD) confidence intervals of Leo et al.104 for the same nodes, we followed their protocol in setting the priors for these nodes. Accordingly, we set the
prior at the root of the tree to have a normal distribution and a mean of 510 Ma with a standard deviation of 7 Ma. The second prior was placed at the split between the higher insects
(_Japyx solifugus_ and _Trigoniophthalmus alternatus_) and Collembola, with a normal distribution and a mean of 485 Ma with a standard deviation of 6 Ma (for the rationale behind this prior
assignment, see Leo et al.104). A Bayesian consensus phylogenetic tree was calculated using BEAST submitted through the CIPRES Science Gateway105 from three independent chains at different
starting seeds for a chain length of 280 million generations, a thinning interval of 10,000, and 25% burn-in to ensure that stationarity was reached. Convergence was assessed using Tracer
v1.7106, trees were summarised using TreeAnnotator v2.6.298 (20% burn-in), and the resulting topology was visualised in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). ANCESTRAL
STATE RECONSTRUCTION A quantitative phyletic approach was used to assess the evolutionary relationships between taxa based on this biological information. Ancestral state reconstructions
were performed using both parsimony and maximum likelihood methods in Mesquite v3.61107; the discrete trait of each taxon (from Supplementary Information Dataset S1) was provided as an input
for the analyses. The likelihood ratio test of the maximum likelihood method was used for the Markov k-state one-parameter (Mk1) and the asymmetrical Markov k-state 2 parameter (AsymmMk)
models107,108, while the rate of character evolution was estimated under the Mk1 model. The Bayesian consensus phylogenetic tree obtained from the 28,000 trees (after the burn-in) sampled
from BEAST was used for the ancestral state reconstruction. The retention index (RI)60 was evaluated for each life form trait to assess phylogenetic conservatism and homoplasy across the
phylogeny. High RI values represent little to no homoplasy (typically above 0.85, see Yu et al.53), while low RI values indicate homoplasy and low phylogenetic signal in a given dataset. To
account for uncertainties with phylogenetic mapping and the occurrence of topographical incongruence across sampled trees, a Bayesian MCMC method implemented in BayesTraits v3.0.2109, was
used for ancestral state reconstructions. The _AddTag _and_ AddMRCA_ options were implemented to find the ancestral state associated with the node of collembolan clade. These options were
used to overcome uncertainty when estimating ancestral states109. A reversible-jump (RJ) hyper-prior was implemented to reduce uncertainty and arbitrariness when selecting priors109. The
hyper-prior draws up values from a uniform distribution between 0 and 30 which is used to seed the exponential distribution for the MCMC analyses. Three independent MCMC chains were run for
30 million iterations with a 25% burn-in. These analyses were repeated with a gamma reversible-jump hyper-prior (mean: 0, 30; variance 0, 30), but no appreciable difference was identified
between the posterior distributions between these analyses when visualised in Tracer. MCMC convergence was assessed in Tracer by ensuring that the effective sample size (ESS) ≥ 200 for all
estimated parameters. The proportion of each state was calculated from the mean values from approximately 20,000 trees randomly chosen by BayesTraits from the 28,000 trees sampled from
BEAST. The states that were reconstructed to be equivocal (probability of 0.2 for each state) were discarded and the probability that the collembolan clade was in a given state was
determined. The average of the estimated probability for the three independent runs was calculated, and any character state with a proportion of > 0.7 was considered to be supported110.
SELECTION ANALYSIS The ratio of nonsynonymous, (dN; i.e. amino acid-altering) to synonymous (dS; i.e. silent) substitution rates, ω = dN/dS, is used to understand the evolutionary dynamics
of genes61,62,63,64. We computed ω using the codon-based maximum likelihood (CodeML) algorithm in EasyCodeML v1.2162,111. To evaluate the influence of selection on the PCG sequence
alignments, we implemented the preset (nested models) running mode with the site model within the framework of the topology of the phylogeny retrieved by the phylogenetic analyses above112.
The codon substitution models described by Geo et al.62, and the significance of their respective dN/dS ratios, were evaluated using the likelihood ratio test (LRT). To identify whether a
link between signatures of mitochondrial selection and a shift in ecological life form exists, we determined the number of nonsynonymous mutations per gene as a proportion of the total
mutations (i.e. dN/(dN + dS)) for all terminal branches, and then averaged these proportions per branch. Moreover, we compared the proportion of nonsynonymous mutations for each life form
shift (aquatic, myrmecophilous, euedaphic, and epiedaphic) vs. the respective sister branches/taxa that represented the ancestral state (hemiedaphic) to ascertain whether a link between the
number nonsynonymous mutations to the ecological life form transition exists. Where multiple branches were present (clades), we accounted for the number of taxa by averaging the proportion
of mutations for each terminal branch, and then followed the path from the terminal branch back until reaching the node where life form divergence (i.e. nearest common ancestor) took place.
Statistical analyses were performed using a paired _t_ test to compare the proportion of synonymous to nonsynonymous mutations across the phylogeny as well as where each branch is paired
with its sister branch that represents a life form shift. Plots of regression analyses were performed using the R package ggplot2113 to assess the correlation between the proportions of
nonsynonymous changes vs. branch length for the taxa that retained the ancestral state and for those who represent a life form shift (i.e. the alternative state). Univariate general linear
models of the regressions were performed in SPSS 27 (Inc., Chicago, IL, USA). Furthermore, and following the same method as described above, we assessed whether there was a link between
selection and species that originated from different latitudinal gradients. This was done twice, first by identifying species across latitudinal gradients where their ranges overlapped, and
secondly by selecting species that had no overlapping distributions. A phylogenetic tree was reconstructed for both analyses, and selection analyses were conducted, and the number of
nonsynonymous mutations were determined for each terminal branch/species. Additionally, we sought to determine the evolutionary dynamics in the context of purifying or diversifying selection
for each terminal branch based on the count of nonsynonymous and synonymous mutations (i.e. ω) for each gene and complex. To represent these results to a gene or complex level (in the form
of a percentage), the number of changes per gene/complex were standardised based on gene size (i.e. total number of nonsynonymous changes/(gene size × total number of branches) × 100).
RADICAL CHANGES IN PHYSICOCHEMICAL PROPERTIES OF AMINO ACIDS The impact of positive selection on physical and chemical properties of 31 amino acids was evaluated for each of the 13 PCGs
using TreeSAAP v3.2 (Selection on Amino Acid Properties)114 by detecting observed changes inferred by the phylogenetic tree under neutral conditions. The impact of 20 physicochemical
properties (excluding 11 amino acids with < 85% accuracy following McClellan and Ellison115) were evaluated. Under the assumption of neutrality, a significant positive z-score as
calculated by TreeSAAP (z-score > 3.09, _p_ < 0.001) for a physicochemical property is indicative of the prevalence of nonsynonymous substitutions, while a significant negative z-score
(z-score < − 3.09, _p_ < 0.001) indicates the prevalence of synonymous substitutions. A sliding window of 15 codons was utilized, based on a categorical scale of 1 (most conservative)
to 8 (most radical). Only the most radical changes (category scores of 7 and 8, _p_ ≤ 0.001) were considered, and for the physicochemical properties that were significant for both category
7 and 8, only the z-scores that were significant for category 8 were included. DATA AVAILABILITY The authors confirm that all relevant data used for this manuscript are available online from
the GenBank NCBI online repository (https://www.ncbi.nlm.nih.gov/genbank/). All sequence accession numbers are list in the Supplementary Information of this manuscript (Table S1).
REFERENCES * Zachos, F. E. (New) Species concepts, species delimitation and the inherent limitations of taxonomy. _J. Genet._ 97, 811–815. https://doi.org/10.1007/s12041-018-0965-1 (2018).
Article PubMed Google Scholar * Zachos, F. E. _et al._ Species inflation and taxonomic artefacts—A critical comment on recent trends in mammalian classification. _Mamm. Biol._ 78, 1–6.
https://doi.org/10.1016/j.mambio.2012.07.083 (2013). Article Google Scholar * De Queiroz, K. Species concepts and species delimitation. _Syst. Bot._ 56, 879–886.
https://doi.org/10.1080/10635150701701083 (2007). Article Google Scholar * Morard, R. _et al._ Nomenclature for the nameless: A proposal for an integrative molecular taxonomy of cryptic
diversity exemplified by planktonic Foraminifera. _Syst. Biol._ 65, 925–940. https://doi.org/10.1093/sysbio/syw031 (2016). Article PubMed Google Scholar * Shtolz, N. & Mishmar, D. The
mitochondrial genome—On selective constraints and signatures at the organism, cell, and single mitochondrion levels. _Front. Ecol. Evol._ 7, 342. https://doi.org/10.3389/fevo.2019.00342
(2019). Article Google Scholar * Pease, J. B., Haak, D. C., Hahn, M. W. & Moyle, L. C. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. _PLoS Biol._
14, e1002379. https://doi.org/10.1371/journal.pbio.1002379 (2016). Article CAS PubMed PubMed Central Google Scholar * Rosenblum, E. B., Römpler, H., Schöneberg, T. & Hoekstra, H. E.
Molecular and functional basis of phenotypic convergence in white lizards at White Sands. _Proc. Natl. Acad. Sci. U.S.A._ 107, 2113–2117. https://doi.org/10.1073/pnas.0911042107 (2010).
Article ADS PubMed Google Scholar * Rosenblum, E. B., Parent, C. E. & Brandt, E. E. The molecular basis of phenotypic convergence. _Annu. Ecol. Evol. Syst._ 45, 203–226.
https://doi.org/10.1146/annurev-ecolsys-120213-091851 (2014). Article Google Scholar * Brawand, D. _et al._ The genomic substrate for adaptive radiation in African cichlid fish. _Nature_
513, 375–381. https://doi.org/10.1038/nature13726 (2014). Article ADS CAS PubMed PubMed Central Google Scholar * Dunn, C. W. & Munro, C. Comparative genomics and the diversity of
life. _Zool. Scr._ 45, 5–13. https://doi.org/10.1111/zsc.12211 (2016). Article Google Scholar * Finch, T. M., Zhao, N., Korkin, D., Frederick, K. H. & Eggert, L. S. Evidence of
positive selection in mitochondrial complexes I and V of the African elephant. _PLoS One_ 9, e92587. https://doi.org/10.1371/journal.pone.0092587 (2014). Article ADS CAS PubMed PubMed
Central Google Scholar * van den Heuvel, L. P. & Smeitink, J. A. M. Nuclear DNA and oxidative phosphorylation. In _Oxidative Phosphorylation in Health and Disease_ (eds Smeitink, J. A.
M. _et al._) 117–129 (Springer, 2004). https://doi.org/10.1007/0-387-26992-4_7. Chapter Google Scholar * da Fonseca, R. R., Johnson, W. E., O’Brien, S. J., Ramos, M. J. & Antunes, A.
The adaptive evolution of the mammalian mitochondrial genome. _BMC Genomics_ 9, 1–22. https://doi.org/10.1186/1471-2164-9-119 (2008). Article CAS Google Scholar * Teacher, A. G. F.,
André, C., Merilä, J. & Wheat, C. W. Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. _BMC Evol. Biol._ 12, 1–14.
https://doi.org/10.1186/1471-2148-12-248 (2012). Article Google Scholar * Pesole, G., Gissi, C., De Chirico, A. & Saccone, C. Nucleotide substitution rate of mammalian mitochondrial
genomes. _J. Mol. Evol._ 48, 427–434. https://doi.org/10.1007/pl00006487 (1999). Article ADS CAS PubMed Google Scholar * Ballard, J. W. O. & Whitlock, M. C. The incomplete natural
history of mitochondria. _Mol. Ecol._ 13, 729–744. https://doi.org/10.1046/j.1365-294x.2003.02063.x (2004). Article PubMed Google Scholar * Galtier, N., Nabholz, B., Glemin, S. &
Hurst, G. D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. _Mol. Ecol._ 18, 4541–4550. https://doi.org/10.1111/j.1365-294x.2009.04380.x (2009). Article CAS PubMed
Google Scholar * Moritz, C. Applications of mitochondrial DNA analysis in conservation: A critical review. _Mol. Ecol._ 3, 401–411. https://doi.org/10.1111/j.1365-294x.1994.tb00080.x
(1994). Article CAS Google Scholar * Avise, J. C. & Ellis, D. Mitochondrial DNA and the evolutionary genetics of higher animals. _Philos. Trans. R. Soc. B Biol. Sci._ 312, 325–342.
https://doi.org/10.1098/rstb.1986.0011 (1986). Article ADS CAS Google Scholar * Kimura, M. _The Neutral Theory of Molecular Evolution_ (Cambridge University Press, 1983). Book Google
Scholar * Kimura, M. The neutral theory of molecular evolution: A review of recent evidence. _J. Jpn. Genet._ 66, 367–386. https://doi.org/10.1266/jjg.66.367 (1991). Article CAS Google
Scholar * Ballard, J. W. O. & Kreitman, M. Is mitochondrial DNA a strictly neutral marker?. _Trends Ecol. Evol._ 10, 485–488. https://doi.org/10.1016/s0169-5347(00)89195-8 (1995).
Article CAS PubMed Google Scholar * Rand, D. M. The units of selection on mitchondrial DNA. _Annu. Rev. Ecol. Syst._ 32, 415–448. https://doi.org/10.1146/annurev.ecolsys.32.081501.114109
(2001). Article Google Scholar * Ballard, J. W. O. & Kreitman, M. Unraveling selection in the mitochondrial genome of _Drosophila_. _Genetics_ 138, 757–772.
https://doi.org/10.1093/genetics/138.3.757 (1994). Article CAS PubMed PubMed Central Google Scholar * Ballard, J. W. O. & Rand, D. M. The population biology of mitochondrial DNA and
its phylogenetic implications. _Annu. Ecol. Evol. Syst._ 36, 621–642. https://doi.org/10.1146/annurev.ecolsys.36.091704.175513 (2005). Article Google Scholar * Teske, P. R. _et al._
Mitochondrial DNA is unsuitable to test for isolation by distance. _Sci. Rep._ 8, 1–9. https://doi.org/10.1038/s41598-018-25138-9 (2018). Article CAS Google Scholar * Shen, Y.-Y. _et al._
Adaptive evolution of energy metabolism genes and the origin of flight in bats. _Proc. Natl. Acad. Sci._ 107, 8666–8671. https://doi.org/10.1073/pnas.0912613107 (2010). Article ADS PubMed
PubMed Central Google Scholar * Yu, L., Wang, X., Ting, N. & Zhang, Y. Mitogenomic analysis of Chinese snub-nosed monkeys: Evidence of positive selection in NADH dehydrogenase genes
in high-altitude adaptation. _Mitochondrion_ 11, 497–503. https://doi.org/10.1016/j.mito.2011.01.004 (2011). Article CAS PubMed Google Scholar * Silva, G., Lima, F. P., Martel, P. &
Castilho, R. Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. _Proc. R. Soc. B_ 281, 20141093. https://doi.org/10.1098/rspb.2014.1093 (2014). Article CAS
PubMed PubMed Central Google Scholar * Consuegra, S., John, E., Verspoor, E. & de Leaniz, C. G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted
fish species. _Genet. Sel. Evol._ 47, 1–10. https://doi.org/10.1186/s12711-015-0138-0 (2015). Article CAS Google Scholar * Li, X. _et al._ Positive selection drove the adaptation of
mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. _Front. Genet._ 9, 605. https://doi.org/10.3389/fgene.2018.00605 (2018). Article CAS PubMed
PubMed Central Google Scholar * Lajbner, Z., Pnini, R., Camus, M. F., Miller, J. & Dowling, D. K. Experimental evidence that thermal selection shapes mitochondrial genome evolution.
_Sci. Rep._ 8, 1–12. https://doi.org/10.1038/s41598-018-27805-3 (2018). Article CAS Google Scholar * Briones, M. J. I. The serendipitous value of soil fauna in ecosystem functioning: The
unexplained explained. _Front. Environ. Sci._ 6, 149. https://doi.org/10.3389/fenvs.2018.00149 (2018). Article Google Scholar * Orgiazzi, A. _et al._ Geographical and temporal
distribution. In _Global Soil Biodiversity Atlas_ (eds Orgiazzi, A. _et al._) 66–91 (European Commission, Publications Office of the European Union, 2016). https://doi.org/10.2788/2613.
Chapter Google Scholar * Bardgett, R. D., Yeates, G. W. & Anderson, J. M. Patterns and determinants of soil biological diversity. In _Biological Diversity and Functions in Soils_ (eds
Bardgett, R. D. _et al._) 100–118 (Cambridge University Press, 2005). https://doi.org/10.1017/cbo9780511541926.007. Chapter Google Scholar * D’Haese, C. A. Were the first springtails
semi-aquatic? A phylogenetic approach by means of 28S rDNA and optimization alignment. _Proc. R. Soc. Lond. B_ 269, 1143–1151. https://doi.org/10.1098/rspb.2002.1981 (2002). Article CAS
Google Scholar * Kristensen, N. P. Phylogeny of insect orders. _Annu. Rev. Entomol._ 26, 135–157. https://doi.org/10.1146/annurev.en.26.010181.001031 (1981). Article Google Scholar *
Carapelli, A., Nardi, F., Dallai, R. & Frati, F. A review of molecular data for the phylogeny of basal hexapods. _Pediobiologia_ 50, 191–204. https://doi.org/10.1016/j.pedobi.2006.01.001
(2006). Article CAS Google Scholar * Nardi, F. _et al._ Hexapod origins: Monophyletic or paraphyletic?. _Science_ 299, 1887–1889. https://doi.org/10.1126/science.1078607 (2003). Article
ADS CAS PubMed Google Scholar * Cicconardi, F., Fanciulli, P. P. & Emerson, B. C. Collembola, the biological species concept and the underestimation of global species richness.
_Mol. Ecol._ 22, 5382–5396. https://doi.org/10.1111/mec.12472 (2013). Article CAS PubMed Google Scholar * Whalley, P. & Jarzembowski, E. A. A new assessment of _Rhyniella_, the
earliest known insect, from the Devonian of Rhynie, Scotland. _Nature_ 291, 317. https://doi.org/10.1038/291317a0 (1981). Article ADS Google Scholar * Hirst, S. & Maulik, S. On some
arthropod remains from the Rhynie Chert (old red sandstone). _Geol. Mag._ 63, 69–71. https://doi.org/10.1017/s0016756800083692 (1926). Article ADS Google Scholar * Deharveng, L., D’Haese,
C. A. & Bedos, A. Global diversity of springtails (Collembola; Hexapoda) in freshwater. _Hydrobiologia_ 595, 329–338. https://doi.org/10.1007/s10750-007-9116-z (2008). Article Google
Scholar * Hopkin, S. P. _Biology of the Springtails (Insecta: Collembola)_ (Oxford University Press, 1997). Google Scholar * Ponge, J., Dubs, F., Gillet, S., Sousa, J. P. & Lavelle, P.
Decreased biodiversity in soil springtail communities: The importance of dispersal and landuse history in heterogeneous landscapes. _Soil Biol. Biochem._ 38, 1158–1161.
https://doi.org/10.1016/j.soilbio.2005.09.004 (2006). Article CAS Google Scholar * Auclerc, A., Ponge, J. F., Barot, S. & Dubs, F. Experimental assessment of habitat preference and
dispersal ability of soil springtails. _Soil Biol. Biochem._ 41, 1596–1604. https://doi.org/10.1016/j.soilbio.2009.04.017 (2009). Article CAS Google Scholar * Salmon, S. _et al._ Linking
species, traits and habitat characteristics of Collembola at European scale. _Soil Biol. Biochem._ 75, 73–85. https://doi.org/10.1016/j.soilbio.2014.04.002 (2014). Article CAS Google
Scholar * Rusek, J. Biodiversity of Collembola and their functional role in the ecosystem. _Biodivers. Conserv._ 7, 1207–1219. https://doi.org/10.1023/a:1008887817883 (1998). Article
Google Scholar * Potapov, A. A., Semenina, E. E., Korotkevich, A. Y., Kuznetsova, N. A. & Tiunov, A. V. Connecting taxonomy and ecology: Trophic niches of collembolans as related to
taxonomic identity and life forms. _Soil Biol. Biochem._ 101, 20–31. https://doi.org/10.1016/j.soilbio.2016.07.002 (2016). Article CAS Google Scholar * Malcicka, M., Berg, M. P. &
Ellers, J. Ecomorphological adaptations in Collembola in relation to feeding strategies and microhabitat. _Eur. J. Soil Biol._ 78, 82–91. https://doi.org/10.1016/j.ejsobi.2016.12.004 (2017).
Article Google Scholar * Parisi, V., Menta, C., Gardi, C., Jacomini, C. & Mozzanica, E. Microarthropod communities as a tool to assess soil quality and biodiversity: A new approach in
Italy. _Agric. Ecosyst. Environ._ 105, 323–333. https://doi.org/10.1016/j.agee.2004.02.002 (2005). Article Google Scholar * Martins da Silva, P. _et al._ Traits of collembolan life-form
indicate land use types and soil properties across an European transect. _Appl. Soil Ecol._ 97, 69–77. https://doi.org/10.1016/j.apsoil.2015.07.018 (2016). Article Google Scholar * Yu, D.
_et al._ Molecular phylogeny and trait evolution in an ancient terrestrial arthropod lineage: Systematic revision and implications for ecological divergence (Collembola, Tomocerinae). _Mol.
Phylogenet. Evol._ 154, 106995. https://doi.org/10.1016/j.ympev.2020.106995 (2020). Article PubMed Google Scholar * Robin, N., D’Haese, C. & Barden, P. Fossil amber reveals
springtails’ longstanding dispersal by social insects. _BMC Evol. Biol._ 19, 1–12. https://doi.org/10.1186/s12862-019-1529-6 (2019). Article CAS Google Scholar * Cicconardi, F., Nardi,
F., Emerson, B. C., Frati, F. & Fanciulli, P. P. Deep phylogeographic divisions and long-term persistence of forest invertebrates (Hexapoda: Collembola) in the North-Western
Mediterranean basin. _Mol. Ecol._ 19, 386–400. https://doi.org/10.1111/j.1365-294x.2009.04457.x (2010). Article CAS PubMed Google Scholar * Deharveng, L. Recent advances in Collembola
systematics. _Pediobiologia_ 48, 415–433. https://doi.org/10.1016/j.pedobi.2004.08.001 (2004). Article Google Scholar * Berg, M. P., Kniese, J., Bedaux, J. & Verhoef, H. A. Dynamics
and stratification of functional groups of micro- and mesoarthropods in the organic layer of a Scots pine forest. _Biol. Fertil. Soils_ 26, 268–284. https://doi.org/10.1007/s003740050378
(1998). Article Google Scholar * Petersen, H. Population dynamic and metabolic characterization of Collembola species in a beech forest ecosystem. In _Soil Biology as Related to Land Use
Practices_ (ed. Dindal, D. L.) 806–833 (Office of Pesticide and Toxic Substance, 1980). Google Scholar * Coulibaly, S. F. M. _et al._ Functional assemblages of Collembola determine soil
microbial communities and associated functions. _Front. Environ. Sci._ 7, 52. https://doi.org/10.3389/fenvs.2019.00052 (2019). Article Google Scholar * Farris, S. The retention index and
the rescaled consistency index. _Cladistics_ 5, 417–419. https://doi.org/10.1111/j.1096-0031.1989.tb00573.x (1989). Article PubMed Google Scholar * Yang, Z. & Bielawski, J. P.
Statistical methods for detecting molecular adaptation. _Trends Ecol. Evol._ 15, 496–503. https://doi.org/10.1016/s0169-5347(00)01994-7 (2000). Article CAS PubMed PubMed Central Google
Scholar * Gao, F. _et al._ EasyCodeML: A visual tool for analysis of selection using CodeML. _Ecol. Evol._ 9, 3891–3898. https://doi.org/10.1002/ece3.5015 (2019). Article PubMed PubMed
Central Google Scholar * Ina, Y. Pattern of synonymous and nonsynonymous substitutions: An indicator of mechanisms of molecular evolution. _J. Genet._ 75, 91–115.
https://doi.org/10.1007/bf02931754 (1996). Article CAS Google Scholar * Yang, Z. & Nielsen, R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. _J. Mol. Evol._
46, 409–418. https://doi.org/10.1007/pl00006320 (1998). Article ADS CAS PubMed Google Scholar * Choudhuri, S. Fundamentals of molecular evolution. In _Bioinformatics for Beginners:
Genes, Genomes, Molecular Evolution, Databases and Analytical Tools_ (ed. Choudhuri, S.) 27–53 (Academic Press, 2014). https://doi.org/10.1016/b978-0-12-410471-6.00002-5. Chapter Google
Scholar * George, D. & Blieck, A. Rise of the earliest tetrapods: An early Devonian origin from marine environment. _PLoS One_ 6, e22136. https://doi.org/10.1371/journal.pone.0022136
(2011). Article ADS CAS PubMed PubMed Central Google Scholar * Zhang, F. & Deharveng, L. Systematic revision of Entomobryidae (Collembola) by integrating molecular and new
morphological evidence. _Zool. Scr._ 44, 298–311. https://doi.org/10.1111/zsc.12100 (2014). Article Google Scholar * Zhang, F., Sun, D., Yu, D. & Wang, B. Molecular phylogeny supports
S-chaetae as a key character better than jumping organs and body scales in classification of Entomobryoidea (Collembola). _Sci. Rep._ 5, 12471. https://doi.org/10.1038/srep12471 (2015).
Article ADS PubMed PubMed Central Google Scholar * Chen, T., Sandmann, P., Schaefer, I. & Scheu, S. Neutral lipid fatty acid composition as trait and constraint in Collembola
evolution. _Ecol. Evol._ 7, 9624–9638. https://doi.org/10.1002/ece3.3472 (2017). Article PubMed PubMed Central Google Scholar * Ding, Y., Yu, D., Guo, W., Li, J. & Zhang, F.
Molecular phylogeny of Entomobrya (Collembola: Entomobryidae) from China: Colour pattern groups and multiple origins. _Insect Sci._ 26, 587–597. https://doi.org/10.1111/1744-7917.12559
(2018). Article PubMed Google Scholar * Zhang, F. _et al._ Molecular phylogeny reveals independent origins of body scales in Entomobryidae (Hexapoda: Collembola). _Mol. Phylogenet. Evol._
70, 231–239. https://doi.org/10.1016/j.ympev.2013.09.024 (2014). Article PubMed Google Scholar * Timmermans, M. J. T. N., Roelofs, D., Mariën, J. & van Straalen, N. M. Revealing
pancrustacean relationships: Phylogenetic analysis of ribosomal protein genes places Collembola (springtails) in a monophyletic Hexapoda and reinforces the discrepancy between mitochondrial
and nuclear DNA markers. _BMC Evol. Biol._ 8, 83. https://doi.org/10.1186/1471-2148-8-83 (2008). Article CAS PubMed PubMed Central Google Scholar * Misof, B. _et al._ Phylogenomics
resolves the timing and pattern of insect evolution. _Science_ 346, 763–767. https://doi.org/10.1126/science.1257570 (2014). Article ADS CAS PubMed Google Scholar * Beutel, R. G.,
Yavorskaya, M. I., Mashimo, Y. & Fukui, M. The phylogeny of Hexapoda (Arthropoda) and the evolution of megadiversity. _Proc. Arthropodan Embryol. Soc. Jpn._ 51, 1–15 (2017). Google
Scholar * Carapelli, A., Liò, P., Nardi, F., Van der Wath, E. & Frati, F. Phylogenetic analysis of mitochondrial protein coding genes confirms the reciprocal paraphyly of Hexapoda and
Crustacea. _BMC Evol. Biol._ 7, 1–13. https://doi.org/10.1186/1471-2148-7-s2-s8 (2007). Article Google Scholar * D’Haese, C. A. Morphological appraisal of Collembola phylogeny with special
emphasis on Poduromorpha and a test of the aquatic. _Zool. Scr._ 32, 563–586. https://doi.org/10.1046/j.1463-6409.2003.00134.x (2003). Article Google Scholar * Magallón, S. A review of
the effect of relaxed clock method, long branches, genes, and calibrations in the estimation of angiosperm age. _Bot. Sci._ 92, 1–22. https://doi.org/10.17129/botsci.37 (2014). Article
Google Scholar * Pimentel, M., Escudero, M., Sahuquillo, E., Minaya, M. Á. & Catalán, P. Are diversification rates and chromosome evolution in the temperate grasses (Pooideae)
associated with major environmental changes in the Oligocene-Miocene?. _PeerJ_ 5, 1–29. https://doi.org/10.7717/peerj.3815 (2017). Article CAS Google Scholar * Estep, M. C. _et al._
Allopolyploidy, diversification, and the Miocene grassland expansion. _PNAS_ 111, 15149–15154. https://doi.org/10.1073/pnas.1404177111 (2014). Article ADS CAS PubMed PubMed Central
Google Scholar * Strömberg, C. Decoupled taxonomic radiation and ecological expansion of open-habitat grasses in the Cenozoic of North America. _PNAS_ 102, 11980–11984.
https://doi.org/10.1073/pnas.0505700102 (2005). Article ADS CAS PubMed PubMed Central Google Scholar * Fjellberg, A. _The Collembola of Fennoscandia and Denmark, Part II:
Entomobryomorpha and Symphypleona_ (Koninklijke Brill NV, 2007). Book Google Scholar * Moreau, C. S., Bell, C., Vila, R., Archibald, S. & Pierce, N. Phylogeny of the ants:
Diversification in the age of angiosperms. _Science_ 312, 101–104. https://doi.org/10.1126/science.1124891 (2006). Article ADS CAS PubMed Google Scholar * Peters, R. S. _et al._
Evolutionary history of the Hymenoptera. _Curr. Biol._ 27, 1013–1018. https://doi.org/10.1016/j.cub.2017.01.027 (2017). Article CAS PubMed Google Scholar * Prinzing, A. & Woas, S.
Habitat use and stratification of Collembola and oribatid mites. In _Arthropods of Tropical Forests: Spatio-temporal Dynamics and Resource Use in the Canopy_ (eds Basset, Y. _et al._)
271–281 (Cambridge University Press, 2003). Google Scholar * Satta, Y., Ishiwa, H. & Chigusa, S. I. Analysis of nucleotide substitutions of mitochondrial DNAs in _Drosophila
melanogaster_ and its sibling species. _Mol. Biol. Evol._ 4, 638–650. https://doi.org/10.1093/oxfordjournals.molbev.a040464 (1987). Article CAS PubMed Google Scholar * Moriyama, E. N.
& Powell, J. R. Synonymous substitution rates in _Drosophila_: Mitochondrial versus nuclear genes. _J. Mol. Evol._ 45, 378–391. https://doi.org/10.1007/pl00006243 (1997). Article ADS
CAS PubMed Google Scholar * Lawrie, D. S., Messer, P. W., Hershberg, R. & Petrov, D. A. Strong purifying selection at synonymous sites in _D. melanogaster_. _PLoS Genet._ 9, 33–40.
https://doi.org/10.1371/journal.pgen.1003527 (2013). Article CAS Google Scholar * Nachman, M. W. Deleterious mutations in animal mitochondrial DNA. _Genetica_ 102(103), 61–69.
https://doi.org/10.1007/978-94-011-5210-5_6 (1998). Article PubMed Google Scholar * Meiklejohn, C. D., Montooth, K. L. & Rand, D. M. Positive and negative selection on the
mitochondrial genome. _Trends Genet._ 23, 259–263. https://doi.org/10.1016/j.tig.2007.03.008 (2007). Article CAS PubMed Google Scholar * Stewart, J. B. _et al._ Strong purifying
selection in transmission of mammalian mitochondrial DNA. _PLoS Biol._ 6, 63–71. https://doi.org/10.1371/journal.pbio.0060010 (2008). Article CAS Google Scholar * Ponnuswamy, P. K.,
Prabhakaran, M. & Manavalan, P. Hydrophobic packing and spatial arrangement of amino acid residues in globular proteins. _Biochim. Biophys. Acta_ 623, 301–316.
https://doi.org/10.1016/0005-2795(80)90258-5 (1980). Article CAS PubMed Google Scholar * Gromiha, M. M., Oobatake, M., Kono, H., Uedaira, H. & Sarai, A. Relationship between amino
acid properties and protein stability: Buried mutations. _J. Protein Chem._ 18, 565–578. https://doi.org/10.1023/a:1020603401001 (1999). Article CAS PubMed Google Scholar * Kumar, S.,
Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. _Mol. Biol. Evol._ 35, 1547–1549.
https://doi.org/10.1093/molbev/msy096 (2018). Article CAS PubMed PubMed Central Google Scholar * Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and
evolution. _Mol. Biol. Evol._ 35, 1550–1552. https://doi.org/10.1093/molbev/msy073 (2018). Article CAS PubMed PubMed Central Google Scholar * Castoe, T. A., Jiang, Z. J., Gu, W., Wang,
Z. O. & Pollock, D. D. Adaptive evolution and functional redesign of core metabolic proteins in snakes. _PLoS One_ 3, e2201. https://doi.org/10.1371/journal.pone.0002201 (2008). Article
ADS CAS PubMed PubMed Central Google Scholar * Xia, X. & Lemey, P. Assesing substitution saturation with DAMBE. In _Phylogenetic Handbook: A Practical Approach to Phylogenetic
Analysis and Hypothesis Testing_ (eds Lemey, P. _et al._) 611–626 (Cambridge University Press, 2009). Google Scholar * Xia, X., Hafner, M. S. & Sudman, P. D. On transition bias in
mitochondrial genes of pocket gophers. _J. Mol. Evol._ 43, 32–40. https://doi.org/10.1007/bf02352297 (1996). Article ADS CAS PubMed Google Scholar * Bouckaert, R. _et al._ BEAST 2: A
software platform for Bayesian evolutionary analysis. _PLoS Comput. Biol._ 10, e1003537. https://doi.org/10.1371/journal.pcbi.1003537 (2014). Article CAS PubMed PubMed Central Google
Scholar * Deharveng, L. & Bedos, A. Diversity of terrestrial invertebrates in subterranean habitats. In _Cave Ecology, Ecological Studies_ (eds Moldovan, O. T. _et al._) 107–172
(Springer International Publishing, 2018). Chapter Google Scholar * Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation.
_Bioinformatics_ 23, 127–128. https://doi.org/10.1093/bioinformatics/btl529 (2007). Article CAS PubMed Google Scholar * Xiong, Y., Gao, Y., Yin, W. & Luan, Y. Molecular phylogeny of
Collembola inferred from ribosomal RNA genes. _Mol. Phylogenet. Evol._ 49, 728–735. https://doi.org/10.1016/j.ympev.2008.09.007 (2008). Article CAS PubMed Google Scholar * Sun, X. _et
al._ Phylomitogenomic analyses on collembolan higher taxa with enhanced taxon sampling and discussion on method selection. _PLoS One_ 15, e0230827.
https://doi.org/10.1371/journal.pone.0230827 (2020). Article CAS PubMed PubMed Central Google Scholar * Cucini, C. _et al._ Re-evaluating the internal phylogenetic relationships of
Collembola by means of mitogenome data. _Genes_ 12, 44. https://doi.org/10.3390/genes12010044 (2021). Article CAS Google Scholar * Leo, C., Carapelli, A., Cicconardi, F., Frati, F. &
Nardi, F. Mitochondrial genome diversity in Collembola: Phylogeny, dating and gene order. _Diversity_ 11, 169. https://doi.org/10.3390/d11090169 (2019). Article CAS Google Scholar *
Miller, M. A. _et al._ A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. _Evol. Bioinform._ 11, 43–48. https://doi.org/10.4137/ebo.s21501 (2015). Article CAS
Google Scholar * Rambaut, A., Drummond, A., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in bayesian phylogenetics using Tracer 1.7. _Syst. Biol._ 67, 901–904.
https://doi.org/10.1093/sysbio/syy032 (2018). Article CAS PubMed PubMed Central Google Scholar * Maddison, W. P. & Maddison, D. R. Mesquite: A modular system for evolutionary
analysis. http://www.mesquiteproject.org (2019). * Lewis, P. O. A likelihood approach to estimating phylogeny from discrete morphological character data. _Syst. Biol._ 50, 913–925.
https://doi.org/10.1080/106351501753462876 (2001). Article CAS PubMed Google Scholar * Pagel, M., Meade, A. & Barker, D. Bayesian estimation of ancestral character states on
phylogenies. _Syst. Biol._ 53, 673–684. https://doi.org/10.1080/10635150490522232 (2004). Article PubMed Google Scholar * Jordan, F. M., Gray, R. D., Greenhill, S. J. & Mace, R.
Matrilocal residence is ancestral in Austronesian societies. _Proc. R. Soc. B_ 276, 1957–1964. https://doi.org/10.1098/rspb.2009.0088 (2009). Article PubMed PubMed Central Google Scholar
* Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. _Mol. Biol. Evol._ 24, 1586–1591. https://doi.org/10.1093/molbev/msm088 (2007). Article CAS PubMed Google Scholar *
Yang, Z. & Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. _Mol. Biol. Evol._ 19, 908–917.
https://doi.org/10.1093/oxfordjournals.molbev.a004148 (2002). Article CAS PubMed Google Scholar * Wickham, H. _ggplot2: Elegant Graphics for Data Analysis_ (Springer, 2016). Book Google
Scholar * Woolley, S., Johnson, J., Smith, M. J., Crandall, K. A. & Mcclellan, D. A. TreeSAAP: Selection on amino acid properties using phylogenetic trees. _Bioinform. Appl. Note_ 19,
671–672. https://doi.org/10.1093/bioinformatics/btg043 (2003). Article CAS Google Scholar * McClellan, D. A. & Ellison, D. D. Assessing and improving the accuracy of detecting protein
adaptation with the TreeSAAP analytical software. _Int. J. Bioinform. Res. Appl._ 6, 120–133. https://doi.org/10.1504/ijbra.2010.032116 (2010). Article CAS PubMed Google Scholar
Download references ACKNOWLEDGEMENTS The authors thank the National Research Foundation (NRF) Competitive Programme for Rated Researcher grant to B.v.V (NRF CPRR: grant number 138016), the
University of Johannesburg funding to B.v.V. and GES 4.0 fellowship awarded to D.M.M. and D.C.M., and the South African (NRF)/Russia (RFBR) Joint Science and Technology Research
Collaboration project no. 19-516-60002 and no. 118904 (NRF) awarded to M.P. and C.JS., respectively. The authors wish to thank the analyses platform and bioinformatics support provided by
the Centre for High Performance Computing (CHPC) in Cape Town, South Africa. We appreciate the constructive criticism received by the two anonymous reviewers which improved the manuscript.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Centre for Ecological Genomics and Wildlife Conservation, Department of Zoology, University of Johannesburg, Auckland Park, 2006, South Africa
Daniela M. Monsanto, Devon C. Main, Arsalan Emami-Khoyi, Shilpa P. Parbhu, Peter R. Teske & Bettine Jansen van Vuuren * Department of Biological Sciences, University of Cape Town,
Rondebosch, 7701, South Africa Charlene Janion-Scheepers * Iziko South African Museum, Gardens, Cape Town, 8001, South Africa Charlene Janion-Scheepers * Institut de Systématique, Evolution,
Biodiversité (ISYEB)-UMR 7205-CNRS, UPMC, EPHE, Muséum National d’Histoire Naturelle (MNHN), Sorbonne Université, 75231, Paris, France Louis Deharveng & Anne Bedos * Department of
Zoology and Ecology, Institute of Biology and Chemistry, Moscow State Pedagogical University, Moscow, 119991, Russia Mikhail Potapov * Senckenberg Museum of Natural History, 60325, Görlitz,
Germany Mikhail Potapov * School of Natural Sciences, Macquarie University, Sydney, NSW, 2109, Australia Johannes J. Le Roux Authors * Daniela M. Monsanto View author publications You can
also search for this author inPubMed Google Scholar * Devon C. Main View author publications You can also search for this author inPubMed Google Scholar * Charlene Janion-Scheepers View
author publications You can also search for this author inPubMed Google Scholar * Arsalan Emami-Khoyi View author publications You can also search for this author inPubMed Google Scholar *
Louis Deharveng View author publications You can also search for this author inPubMed Google Scholar * Anne Bedos View author publications You can also search for this author inPubMed Google
Scholar * Mikhail Potapov View author publications You can also search for this author inPubMed Google Scholar * Shilpa P. Parbhu View author publications You can also search for this
author inPubMed Google Scholar * Johannes J. Le Roux View author publications You can also search for this author inPubMed Google Scholar * Peter R. Teske View author publications You can
also search for this author inPubMed Google Scholar * Bettine Jansen van Vuuren View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS D.M.M.,
A.E.-K., and B.v.V. designed the research; D.M.M. and D.C.M. analysed data; D.M.M., C.J.-S., L.D., A.B., M.P., and S.P.P. compiled and corrected the ecological life form Supplementary Table;
all authors contributed to the writing of the manuscript. CORRESPONDING AUTHOR Correspondence to Bettine Jansen van Vuuren. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION 1. SUPPLEMENTARY INFORMATION 2. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0
International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the
source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Monsanto, D.M., Main, D.C., Janion-Scheepers, C. _et al._ Mitogenome selection in
the evolution of key ecological strategies in the ancient hexapod class Collembola. _Sci Rep_ 12, 14810 (2022). https://doi.org/10.1038/s41598-022-18407-1 Download citation * Received: 12
December 2021 * Accepted: 10 August 2022 * Published: 31 August 2022 * DOI: https://doi.org/10.1038/s41598-022-18407-1 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative