Screening of lactic acid bacteria strains isolated from iranian traditional dairy products for gaba production and optimization by response surface methodology
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT A total of 50 lactic acid bacteria (LAB) isolates from Iranian traditional dairy products (Motal and Lighvan cheeses, and artisanal yogurt) were screened for gamma-aminobutyric acid
(GABA) production. Firstly, a rapid colorimetric test was performed to evaluate the glutamate decarboxylase (GAD) activity among the LAB isolates examined. Thin layer chromatography (TLC)
was then performed on selected strains to identify isolates with high/moderate GABA producing capacity, and a GABase micro-titer plate assay was employed to quantify GABA. Finally, two
_Lactococcus_ (_Lac._) _lactis_ strains were selected for GABA production optimization via Response Surface Methodology (RSM) following Central Composite Design (CCD). Forty-one out of the
50 isolates showed GAD activity according to the colorimetric assay. Eight isolates displayed strong GAD activity, while nine showed no activity; low to moderate GAD activity was scored for
all other isolates. GABA production was confirmed by TLC in all isolates with high GAD activity and in four selected among isoaltes with moderate activity. Among the _Lactococcus_ strains
tested, _Lac. lactis_ 311 and _Lac. lactis_ 491 were the strongest GABA producers with amounts of 3.3 and 1.26 mM, respectively. These two strains were subjected to GABA production
optimization applying RSM and CCD on three key variables: Monosodium glutamate concentration (MSG) (between 25 and 150 mM), incubation temperature (between 25 and 37 °C), and pH (between 4.0
and 5.0). Optimal conditions for GABA production by _Lac. lactis_ 311 and _Lac. lactis_ 491 of temperature, pH and MSG concentration were, respectively, 35.4 and 30 °C, pH 4.5 and 4.6, and
MSG concentration of 89 and 147.4 mM, respectively. Under the above conditions, the amount of GABA produced by _Lac. lactis_ 311 and _Lac. lactis_ 491 was 0.395 and 0.179 mg/mL,
respectively. These strains and the optimal culture conditions determined in this study could be used for the biotechnological production of GABA or applied in food fermentations for the
development of naturally GABA-enriched foods. SIMILAR CONTENT BEING VIEWED BY OTHERS OPTIMIZING ALPHA-AMYLASE FROM _BACILLUS AMYLOLIQUEFACIENS_ ON BREAD WASTE FOR EFFECTIVE INDUSTRIAL
WASTEWATER TREATMENT AND TEXTILE DESIZING THROUGH RESPONSE SURFACE METHODOLOGY Article Open access 06 November 2023 A REFINED MEDIUM TO ENHANCE THE ANTIMICROBIAL ACTIVITY OF POSTBIOTIC
PRODUCED BY _LACTIPLANTIBACILLUS PLANTARUM_ RS5 Article Open access 07 April 2021 INDUSTRIAL PRODUCTION AND FUNCTIONAL PROFILING OF PROBIOTIC _PEDIOCOCCUS ACIDILACTICI_ 72 N FOR POTENTIAL
USE AS A SWINE FEED ADDITIVE Article Open access 29 April 2025 INTRODUCTION Lactic acid bacteria (LAB) are among the most essential and common Gram-positive bacteria which are not only
widely distributed in nature and naturally exist in traditional fermented foods, but also extensively involved in industrial food fermentations due to having a reputation of being “Generally
Recognized As Safe” (GRAS). The use of pure cultures of lactic acid bacteria for fermentation of cucumbers, cabbage, olives, and other products has been explored for several decades with
varying degrees of success. However, at present pure cultures are exploited only on a limited commercial scale for these commodities1,2,3,4. LAB also offer special functions that are
considered beneficial, such as antioxidant and antimicrobial activities, as well as the formation of bioactive compounds such as, among others, peptides, organic acids, and short-chain fatty
acids5,6,7,8,9. Because of their significant beneficial effects, there are currently noteworthy researches on LAB and attract many industrial interest10. Gamma-aminobutyric acid (GABA) is
considered as one of the well-known bioactive compounds; it is a four-carbon, non-protein amino acid formed by different organisms including microorganisms, plants, and animals3,11,12. This
amino acid-derived compound serves as the main inhibitory neurotransmitter in the mammalian central nervous system and, as such, it leds to hypotension, contributes to gut-to-brain signaling
and has tranquilizer effects7,13,14,15,16. Several studies show that GABA contributes to other physiological functions, such as regulating of sleeplessness and depression, enhancing the
plasma level of growth hormone, controlling the quorum-sensing systems by acting as a signal molecule between eukaryote cells and pathogens, reducing the inflammation in rheumatoid
arthritis10,17,18,19. As a bioactive compound, the application of GABA varies from pharmaceuticals to functional fermented foods20. Therefore, it is an ever-growing demand for highly
effective GABA biosynthesis, and hence, substantial efforts have been made in this field. Biological methods applying microorganisms are more promising; thus many GABA-enriched products are
obtained by fermentation21,22,23,24,25. Biosynthesis of GABA from glutamate occurs by the action of the glutamate decarboxylase (GAD), a pyridoxal 5-phosphate-dependant enzyme (EC 4.1.1.15)
that is responsible for the conversion of L-glutamate into GABA3,24. The wide distribution of GAD among eukaryotes (plants, animals, and fungal strains) and prokaryotes has been reported by
several studies26,27. However, LAB, besides all beneficial and technological properties, have been introduced as the most promising group of microorganisms in which they are capable of
producing a high level of GABA due to high GAD activity28. The effort to screen for new GABA-producing LAB still attracts attention, though numerous strains have already been isolated and
characterized. Thus, it seems that further research on the isolation and characterization of GABA-producing LAB is demanded to provide novel starters for the nutraceutical and functional
fermented foods industry10. The diversity of new GABA-enriched foods ranges from cereal-based to dairy-fermented products, including sourdough, bread, cheese, fermented tea and vegetables,
traditional Asian fermented foods, and dairy and soy products1,29. As mentioned above, by further screening, the isolation sources should be diversified as much as possible, including
traditional fermented foods to achieve new GABA-producing LAB strains. This leads to a more comprehensive application area and higher flexibility of starter cultures11. According to the
literature, raw milk cheeses have been identified as a valuable source of microbial biodiversity and new LAB strains with health-promoting properties14. Though currently, there are several
different types of researches and industrial interest in the biological examination of the potential of traditional dairy LAB30,31,32. So far, no GABA-producing LAB have been reported from
Iranian traditional fermented foods. Recently Azizi et al. (2017) have claimed that most of the isolates from Motal cheese could be used as starters or adjunct-starters in novel fermented
functional foods. Besides having antimicrobial properties33, the production of bioactive compounds, such as GABA, could potentially substantiate the functionality claimed for such cultures.
Hence, the present study aims to screen for GABA-producing LAB from various Motal and Lighvan cheeses and yogurt samples which are among the preeminent and most appreciated traditional
Iranian dairy products. MATERIALS AND METHODS CHEMICAL AND MEDIA The commercial GABase kit and all chemicals such as NADP + (Nicotinamide adenine dinucleotide phosphate), α-ketoglutarate,
monosodium glutamate (MSG), Triton X-100®, and GABA were purchased from Sigma-Aldrich (St. Louis, MO, USA). M17 and de Man, Rogosa, and Sharpe (MRS) broth media were obtained from Quelab
(Quelab Laboratories Inc., Canada). LAB STRAINS AND GROWTH CONDITIONS A total of 50 LAB isolates previously isolated from Iranian traditional dairy products (Motal cheese33, Lighvan
cheeses31,34, and Iranian yogurt35), identified using molecular techniques and 16S rRNA gene sequencing, were screened for GABA production (Table 1). All examined isolates, including
lactobacilli (_Lactobacillus, Lactiplantibacillus_, and _Levilactobacillus_) (27 isolates), _Lactococcus_ (20 isolates), and _Streptococcus_ (3 isolates), were kept frozen in MRS
(lactobacilli) or M17 (lactococci and streptococci) broth with 15% glycerol at -80 °C. Pre-cultures were made by adding 20 µL of stock culture to 10 mL fresh medium. Lactobacilli strains
were grown in MRS medium, while M17 medium was used for lactococci and _Streptococcus_ (_St._) _thermophilus_ strains. Based on the types, incubations were done at 30 or 37ºC for 24–48 h
under aerobic or anaerobic conditions. QUALIFICATION SCREENING OF LAB ISOLATES DETERMINATION OF GAD ACTIVITY Glutamic acid decarboxylase (GAD) assay is a rapid colorimetric test used to
assess the GABA-producing potential among LAB isolates. Sample preparation for GAD activity determination was done as described by Lacroix et al.36. An overnight grown culture of each
isolate (5 mL) was washed with 0.9% (w/v) NaCl solution and centrifuged at 5000 × g (20 min, 25 ºC). Cells were suspended in 500 µL of GAD reagent solution (pH = 4), which consists of
L-glutamic acid (1 g), Triton X-100 (300 µL), NaCl (90 g), and bromocresol green (0.05 g) in 1 L deionized water. After anaerobic incubation for 4 h at 37ºC, GAD activity was visually
examined by color change. The development of a greenish or blueish color was considered as low or high GAD activity, respectively. However, no color change (yellow) was considered with no
activity36. THIN LAYER CHROMATOGRAPHY (TLC) To identify isolates with high and moderate GABA producing ability, a TLC assay was performed as described by Lee et al.37. Briefly, strains were
inoculated in their respective broth medium supplemented with 0.5% monosodium glutamate (MSG) and incubated at their optimal temperature for 24 h. To test the presence of GABA, supernatants
were collected by centrifugation (8000 × g, 4ºC, 5 min) and then 2 µL of each filtered supernatant was spotted on a TLC plate (Silica gel 60 F254). TLC was conducted using n-butanol:acetic
acid:water ratio of 5:2:2 (v/v/v). The silica gel plate was subsequently developed by spraying with a 2% ninhydrin solution and heating at 105ºC for 5 min. DNA EXTRACTION AND STRAIN TYPING
BY REP-PCR Total genomic DNA was extracted from GABA-producing isolates using a commercial kit (High Pure PCR Template Preparation Kit; Roche, Basel, Switzerland), following the
manufacturer’s instructions. To assess the interspecies diversity between isolates, repetitive extragenic profiling PCR (rep-PCR) was done using BoxA2R (5´-ACGTGGTTTGAAGAGATTTTCG-3´) as
reported by Koeuth et al.38. In brief, reactions were performed in 25 µL volume containing 2 µL purified DNA, 12.5 µL Red master mix (Ampliqon, Odense, Denmark), 9.3 µL Nuclease-free water,
and 1.2 µL of the primer. Thermal conditions were as follows: an initial step of denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation and annealing at 94 °C for 29 s, 40 °C
for 1 min, and at 68 °C for 8 min; with a final extension at 68 °C for 10 min. PCR products were then electrophoresed using 1.5% agarose (Merck, Darmstadt, Germany) gel at 75 V for 90 min.
DNA safe stain (0.75 µL/100 mL) (Sina Clon Bio Science, Tehran, Iran) was used for visualization of the gel and the bands photographed under UV light by Gel Doc System. The similarity of the
patterns was expressed by the Simple Matching (SM) coefficient, and the clustering was performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method, with the
Multivariate Statistical Package (MVSP) software version 3.13d31. GABA DETERMINATION Quantitative measurment of GABA in broth medium was done by a GABase microtiter plate assay as described
by Tsukatani et al.39. Bacterial isolates were grown in MRS broth medium containing 0.5% MSG for 2 days at their optimal growth temperature. The reactive mixture contained 750 mM sodium
sulfate, 10 mM dithiothreitol, 1.4 mM NADP + , 2.0 mM α-ketoglutarate in Tris–HCl buffer (80 mM, pH 9.0) and GABA-T (Gamma-aminobutyrate glutamate aminotransferase) (Sigma-Aldrich). The
mixture was added to each well (in a 96-well microtiter plate) in a final volume of 90 µL. The initial absorbance was read at 340 nm with a microplate reader (BioTek, ELX 808) before adding
a 10 µL sample of a standard GABA solution. The substrate of NADP + turns to NADPH in the presence of GABA and α-ketoglutarate after incubation at 30-37ºC for 1 h. To calculate the GABA
content formed by each isolate, the difference between initial and final absorbances at 340 nm was compared to standard curves of calibrated GABA solutions. OPTIMIZATION OF GABA PRODUCTION
USING RESPONSE SURFACE METHODOLOGY (RSM) Optimization of GABA production in MRS medium and studying the interaction of key parameters were done using response surface methodology (RSM). A
Central Composite Design (CCD) of RSM was applied to optimize the 3 effective variables: MSG concentration (between 25 and 150 mM), incubation temperature (from 25 to 37 °C), and pH
(4.0–5.0). Varying these three independent parameters, a total of 20 experiments for each isolate were designed and performed. The statistical software Design Expert 10.0.7.0 was used for
data analysis12,40. Laboratory-scale fermentations were set up to investigate optimal parameters for GABA production by selected isolates. Bacterial isolates were inoculated into falcon
tubes containing 50 mL MRS broth medium with different concentrations of MSG with initial pH adjustment and incubated at different temperatures for 48 h to reach optimum GABA production.
After 48 h of incubation, samples were withdrawn from the each falcon tube to determine GABA content. STATISTICAL ANALYSIS To analyze the results related to the potential of GABA production
by examined bacteria, Duncan’s one-way analysis of variance (ANOVA) was applied using the SPSS software (ver. 16), while to optimize the conditions of GABA production, RSM with Design Expert
10.0.7.0 software was used. RESULTS SCREENING OF GAD ACTIVITY Total number of LAB isolates and the results of their previous identification31,33,34,35 are presented in Table 1. These
isolates include members of lactobacilli (of the genera _Lactobacillus, Lactiplantibacillus_, and _Levilactobacillus_) (27 isolates), _Lactococcus_ (20 isolates), and _Streptococcus_ (3
isolates). The GAD activity of the 50 isolates from Iranian traditional dairy products are also summarized in Table 1. The cultures of the isolates _Levilactobacillus_ (_Lb._) _brevis_ M2,
M4 and M12; _Streptococcus_ (_St._) _thermophilus_ 43, and _Lac. lactis_ 261, 311, 454, and 491 developed a strong blue color, consistent with a high GAD activity. No color change was
observed for the isolates M3, M6, M7, M8, M9, M13, M15, M17, and M19; thus these are considered not to have GAD activity. Other isolates with greenish-blue, green and greenish-yellow color
were considered as having medium, low and very low GAD activity, respectively. Fourteen isolates, including all high and some with moderate GAD activity were selected for subsequent
experiments. GAD assay results revealed that most of the GAD-positive isolates were isolated from cheese (Lighvan and Motal); whereas, only one isolate from a traditional yogurt exhibited
high GAD activity. GENETIC ANALYSIS OF ISOLATES Lactobacilli, _Streptococcus_ and _Lactococcus_ species were targeted by rep-PCR to evaluate the intramolecular diversity among the isolates,
as well as to assess whether isolates with GAD activity belonging to the same species were replicates or independent strains. Further, typing of the strains could serve for selecting
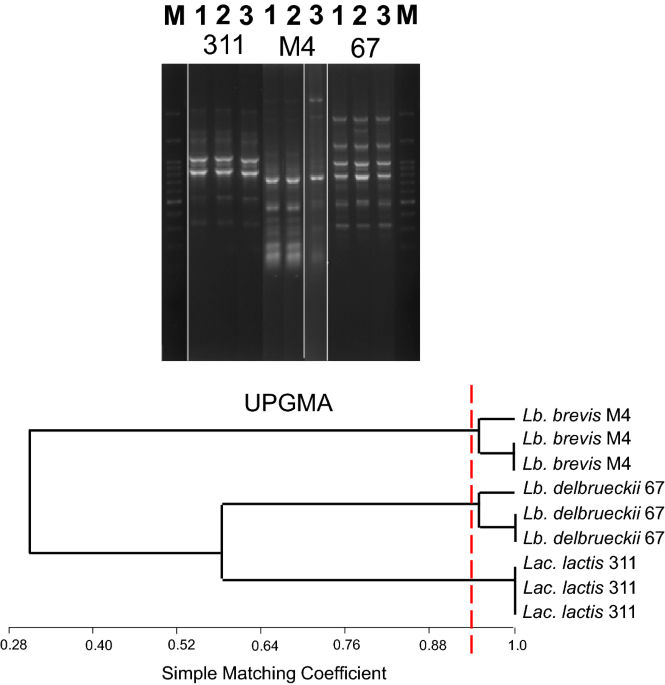
different strains for GABA production optimization. The different patterns of similarity were clustered using the SM coefficient by the UPGMA method. Given the reproducibility of the assay
around 94 (Fig. 1), isolates sharing a percentage of similarity > 94% were considered to belong to the same strain. Figure 2 a, b, c and d are the profiles obtained from 3 isolates of
_Lb. brevis_, 4 isolates of _Lac. lactis_, 3 isolates of _St. thermophilus_ and 4 isolates of _Lb. delbrueckii. Lb. brevis_ M2 and _Lb. brevis_ M12 strains showed 100% similarity. _Lac.
lactis_ 311 and _Lac. lactis_ 454 strains had 100% similarity. _St. thermophilus_ 74 and _St. thermophilus_ 43 also had 100% similarity. These strains can be multiple isolates.
_Lactobacillus_ (_Lb._) _delbrueckii_ 69 and _Lb. delbrueckii_ 67 also showed very high homology. The relation between the intra-species diversity with GABA production in these species
showed that GABA production was significantly different between isolates with high similarity and comparable between isolates with low similarity. Unlike other examined strains, there was no
significant difference in GABA production in _St. thermophilus_ isolates with high and low similarity. CONFIRMATION OF GABA BY THIN LAYER CHROMATOGRAPHY Isolates that showed no GAD activity
by the colorimetric method were excluded from further analyses. Based on the results of GAD activity, we selected 14 isolates corresponding to the species _St. thermophilus_ (three), _Lb.
delbrueckii_ (four), _Lac. lactis_ (four), _Lb. brevis_ (three) for further studies. The GABA production was confirmed by observing red spots on the TLC plates (Fig. 3). The spot mobility
from the culture supernatant of the 14 isolates was consistent with GABA, as a retention factor (Rf) value (0.77 cm) of the sample matched with the GABA standard. Thus, all 14 selected
isolates were considered as GABA producers. GABA’s of strains show the same retention factor (Rf) value as the GABA standard (= 0.77 cm). The retention factor was defined as the ratio of the
distance traveled by the center of a spot to the distance traveled by the solvent front. QUANTITATIVE GABA PRODUCTION MEASUREMENT GABA production of selected strains was quantified using a
GABase microtiter plate enzymatic determination. As shown in Table 1, GABA production varied widely ranging from 0.01 to 0.36 mg/L (equivalent to 0.12 and 3.52 mM). All _Lb. brevis_ isolates
from Motal cheese exhibited a high ability to produce GABA (> 1 mM), while the highest GABA concentration was produced by _Lb. brevis_ M4. Our findings further revealed that all isolates
belonging to _Lac. lactis_ have a medium to high GABA producing ability, while all _Lb. delbrueckii_ cultures presented a low level of this ability. RSM DESIGN Although _Lac. lactis_
isolates produced less GABA than the _Lb. brevis_ isolates, the former species is the most economically-important starter component in cheese43, which prompted us to select two different
strain of this species, _Lac. lactis_ 311 and _Lac. lactis_ 491, for GABA production optimization by RSM methodology. The selected two _Lac. lactis_ strains are different strains according
to the rep-typing results.As stated in the corresponding section, a CCD (central composite design) was performed based on a single variable factor test at a time and considering three
independent variables: MSG concentration (mM), Temperature (°C), and pH. The 20 different combination set ups based on CCD and the corresponding GABA concentration for both predicted and
experimental values are listed in Tables 2 and 3, respectively. The interaction effect of these factors on GABA production concentration by _Lac. lactis_ 311 is described by the following
equation: $$\begin{aligned} {\text{R}} = & 0.{18} + 0.0{23} \times {\text{A}} + 0.0{19} \times {\text{B}} + {5}.{\text{429E}} - 00{3} \times {\text{C}} + {2}.{\text{363E}} - 00{3} \times
{\text{AB}} - {1}.{\text{937E}} \\ - 00{3} \times {\text{AC}} + {3}.{\text{863E}} - 00{3} \times {\text{BC}} - 0.0{16} \times {\text{A}}^{{2}} - 0.0{2}0 \times {\text{B}}^{{2}} - 0.0{13}
\times {\text{C}}^{{2}} \\ \end{aligned}$$ (1) where R is the concentration of GABA (mg/mL) and A, B and C are coded values of the independent variables, viz., MSG, temperature and pH,
respectively. Three-dimensional surface plots were generated to estimate the effect of the combinations of the independent variables on the GABA production using Design Expert software. The
3D plot in Fig. 4A shows the interaction effect of MSG and temperature, keeping pH at its central level 4.5. According to the 3D plot, predictably, the GABA concentration increased with
increasing MSG concentration and the increase of temperature. Nevertheless, in the low MSG concentration, increasing GABA concentration was more sharply compared to a higher concentration.
The maximum concentration (3.83 mM) was observed with MSG in concentration of 89 mg/mL and temperature of 35.4 °C. Figure 4B presents the 3D plot showing the dependency of GABA concentration
on MSG concentration and pH, keeping the third factor (temperature) at its central level of 31 °C. The graph predicted that the highest GABA concentration by relevant strain at 35.4 °C was
observed with the mid pH (4.5) and MSG concentration (89 mg/ml). However, GABA concentration at lower pH remained approximately constant by increasing MSG concentration. The 3D curve showing
the interaction effect of temperature and pH has shown in Fig. 4C. The low GABA concentration was observed with the lowest temperature and lowest pH. The maximum GABA concentration (3.83
mM) was observed with pH (4.5) and temperature (35.4 °C). Equation 2 explains the interaction effect of the independent variables on GABA concentration by _Lac. lactis_ with strain code 491:
$$\begin{aligned} {\text{GABA}} = & 0.{39} + 0.0{32} \times {\text{A}} + 0.0{48} \times {\text{B}} + {6}.{\text{314E}} - 00{3} \times {\text{C}} - 0.0{11} \times {\text{AB}} -
{7}.{\text{459E}} \\ - 00{4} \times {\text{AC}} - 0.0{14} \times {\text{BC}} - 0.0{19} \times {\text{A}}^{{2}} - 0.0{43} \times {\text{B}}^{{2}} - 0.0{28} \times {\text{C}}^{{2}} \\
\end{aligned}$$ (2) Figure 5A showed that with increasing the concentration of monosodium glutamate and increasing the temperature, the amount of GABA formed increased. This trend is in
agreement with the results reported elsewhere by other authors41,42. An increase in the amount of GABA was observed with increasing MSG and pH to 89 mg/ml and 4.59, respectively, at a
constant temperature (Fig. 5B). When the concentration of monosodium glutamate was kept constant, the GABA level increased sharply with increasing temperature, but increased pH led to the
production of low amounts of GABA (Fig. 5C). The incubation temperature is another important factor that may affect GABA production. In addition to the stability and biological activity of
the enzyme, temperature also affects the thermodynamic equilibrium of the reaction. SELECTION OF OPTIMAL TREATMENT AND VALIDATION OF MODEL The results of GABA production by two examined
strains showed the optimal conditions for GABA production by _Lac. lactis_ 311 and _Lac. lactis_ 491 had a temperature of 35.4 and 30 °C, a pH of 4.5, 4.6 and a concentration of 89 and 147.4
mM of MSG, respectively. In the above conditions, the amount of GABA production by these two strains was 3.83 and 1.73 mM, respectively. Examination of the results in Fig. 6 shows that the
points with a good approximation are on the straight line and the values obtained experimentally are in a good agreement with the values predicted by the model and showed the accuracy of the
equation model. DISCUSSION Regarding the Motal cheese isolates in this study, the results are consistent with previously reported levels by Ebadi Nezhad et al. (2020)44. GABA production
begins in the exponential growth phase and increases near to the stationary phase due to an increased in GAD activity, which agrees with GAD being an intracellular enzyme produced in
response to acidic conditions45. The nutrient sources required for the GABA production by bacteria include the primary sources of carbon, nitrogen (including GABA substrate, glutamic acid)
and minerals46. To avoid the use of replicates from the optimization studies, a strain-level typing scheme by rep-PCR was employed. Knowing intra-species diversity also helps to know how
diverse the extend of GABA production is in closely-related and unrelated strains of the same species10. As for many other phenotypic traits, large phenotypic variations have occasionally
been reported for genetically closely-related LAB strains47. Typing of LAB isolates from Koozeh and Lighvan cheese by rep-PCR with BOXA2R primer and typing of GABA-producing LAB isolates
with rep-PCR by (GTG)5 primer has been previously reported by others10,31. As in this work, high intraspecies diversity was reported in the referred studies. Presumptive detection of GABA
production by LAB species by TLC, followed by its confirmative identification and quantification by High-performance liquid chromatography (HPLC) has been previously reported to be a
successful strategy by Zhang et al.48. These authors reported that the mobility spot of the culture supernatant of BC114 strain was basically consistent GABA standard and could be
preliminarily identified as GABA within a negligible margin of error. The measured sample from the fermentation broth (a complex matrix), may also result in incomplete agreement with pure
GABA standards48. The strain under investigation, _Lb. plantarum_ BC114, showed the highest GABA production as measured by HPLC, which reached a value of 1.52 ± 0.07 g/L48. Tanamool et al.49
also used the TLC method to screen GABA-producing lactic acid bacteria strains. These authors found that among 44 LAB, the isolates L10-11 clearly produced the highest GABA based on the TLC
results49. In our results, 14 isolates among 50 LAB were considered as GABA producers. Valenzuela et al.25 showed the ability of _Lb. brevis_ to produce GABA is consistent with other
reported levels for _Lb. brevis_ LMG6906 (0.29 g/L). _Lb. brevis_ CECT8183 isolated from Spanish cheese by Diana et al.17 produced 0.1 g/L of GABA. _Lb. brevis_ BJ20 produced 0.002 g/L in a
fermented sea tangle solution, which is a popular traditional marine food in Korea based on brown seaweed50. _Lb. brevis_ PM17 was among the most potent studied isolates by Franciosi et
al.9. By contrast, no significant GABA production was detected for any _St. thermophilus_ strain isolated from yogurt products, compared to lactobacilli and _Lactococcus_ species in study.
According to the literature, large numbers of strains from these genera from various fermented foods are indicated as GABA-producing LAB13. There are many publications reporting variable
GABA production levels by different LAB species. Among other factors, production could be affected by the GABA detection methods. For instance, _Lb. delbrueckii_ PR1 (63 mg/kg) and _Lac.
lactis_ PU1 showed the highest GABA concentrations9. The results of Ly et al.10 showed _Lb. futsaii_, _Lb. namurensis_ and _Lb. plantarum_ to produce high amounts of GABA in the range of
1.7–2 g/L. Hwang et al.51 have examined GABA production by two _Lac. lactis_ strains in MRS containing 5% MSG at 30 °C. Maximum GABA production was observed after 40 h (1.37 g/L). Redruello
et al.52 studied the effect of 6 _Lac. lactis_ GABA-producing strains in cheese making. GABA accumulated at concentrations up to 0.457 g/Kg in cheese. Galli et al.53 revealed GABA content of
their fermented milks by two mix starters (_Lac. lactis_ and _Lb. rhamnonus_ or _Lb. paracasei_) 0_._185 and 0.319 g/L, respectively by adding 249 mg/L MSG. Santos-Espinosa et al.54
demonstrated the highest GABA concentration (0.086 g/L) in fermented milk with _Lac. lactis_ L-571 at 37 °C with 3 g/L of glutamate substrate. In a study conducted by Tajabadi et al.55 the
optimum conditions for maximum GABA production by _Lb. plantarum_ Taj-Apis362 were an initial glutamic acid concentration of 497.97 mM, culture temperature of 36 °C, initial pH of 5.31 and
incubation time of 60 h, which produced 0.74 g/L of GABA. GABA-producing ability is linked to the activity of GAD enzyme in LAB56. The source of LAB might affect GAD activity and GABA
production capacity, as it was speculated that acidified foods could probably be the natural niche of GABA producers10. Li et al.57 identified and characterized the GABA-producing lactic
acid bacteria and claimed that most LAB strains are able to produce the highest amount of GABA in the range of pH 4 to 5 at 30 to 50 °C and in the presence of glutamic acid. For this reason,
this temperature and pH range was used in their study57. GABA production occurs under acidic conditions. LAB metabolism mainly leads to the production of organic acids such as lactic and
acetic acid. This means that they have to frequently face acidic stress and therefore, they have developed different mechanisms to cope with acidic conditions such as the GAD pathway.
Laroute et al.58 claimed the activation of this pathway occurred after changing the pH to 4.6 and during the stationary phase. Their results are consistent with our results which optimum pH
for GABA production in two examined isolates is 4.5 and 4.6. As reported by Laroute et al.58 a dose of 152 mM glutamate, increased the production of GABA by _Lac. lactis_ NCDO 2118_._ This
dosage of glutamate is similarly optimum dose for GABA production in our examined isolate (_Lac. lactis_ 491). The findings of our work showed _Lac. lactis_ 311 produced more GABA in lower
concentration than _Lac. lactis_ 491. The probable reason that lower concentrations of MSG can increase the efficiency of GABA production in comparison to higher concentrations of it, is
that large amounts of MSG increase the osmotic pressure of cells and disrupt bacterial metabolism, leading to a decrease in GABA efficiency by bacteria 59. Although the optimal
concentrations of MSG are different for various microorganisms in GABA production, some researchers have proven that excessive MSG could inhibit cell growth and minimize GABA
production60,61. Lu et al.62 showed that the highest concentration of GABA production by _Lac. lactis_ isolated from kimchi was at 34° C (3.68 g/L). These results are similar to our results
about _Lac. lactis_ 311. However, they stated that the optimal pH of GABA production was in the range of 7–862, which was not in compliance with the results obtained in our study.
CONCLUSIONS In the present study, firstly, LAB isolates were screened for GABA production according to a qualitative method based on a colorimetric assay. Significant variability in GABA
production was encountered among the different species and strains tested. GABA-producing strains were then subjected to a TLC analysis to detect strains with high GABA producing ability.
Response surface methodology was finally used to optimize conditions for maximum GABA production in two _Lac. lactis_ strains, taking into account the variables affecting the response. The
selected strains and the culture conditions producing the highest amounts of this bioactive compound could be used to develop GABA-enriched functional foods or in other biotechnological
applications. DATA AVAILABILITY The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. ABBREVIATIONS * LAB: Lactic acid
bacteria * GRAS: Generally recognized as safe * GABA: Gamma-aminobutyric acid * GAD: Glutamate decarboxylase * NADP: Nicotinamide adenine dinucleotide phosphate * MSG: Monosodium glutamate
* TLC: Thin layer chromatography * rep-PCR: Repetitive extra genic profiling PCR * RSM: Response surface methodology * CCD: Central composite design * Rf: Retention factor REFERENCES * FDA.
GRAS Exemption Claim for gamma-Aminobutyric acid (GABA) for Use as a Food Ingredient in the United States (U.S.). . (FDA), Food and Drug Administration Co, Pharma Food International; 2015. *
Fleming HP, McFeeters RF, Daeschel MA. _The lactobacilli, pediococci, and leuconostocs: vegetable products_. In, Bacterial Starter Cultures for Foods: 97–118 (CRC Press, 2018). * Yogeswara,
A. I., Kusumawati, I. G., Sumadewi, N. L., Rahayu, E. S. & Indrati, R. Isolation and identification of lactic acid bacteria from Indonesian fermented foods as γ-aminobutyric
acid-producing bacteria. _Int. Food Res. J._ 25(4), 1753–1757 (2018). Google Scholar * Vinderola, G., Ouwehand, A., Salminen, S., & von Wright, A. (Eds.). Lactic acid bacteria:
microbiological and functional aspects. CRC Press (2019). * Abushelaibi, A., Al-Mahadin, S., El-Tarabily, K., Shah, N. P. & Ayyash, M. Characterization of potential probiotic lactic acid
bacteria isolated from camel milk. _LWT - Food Sci. Technol_ 79, 316–325 (2017). Article CAS Google Scholar * Silva, M. S. _et al._ Probiotic properties of Weissella cibaria and
Leuconostoc citreum isolated from tejuino–A typical Mexican beverage. _LWT - Food Sci. Technol._ 86, 227–232 (2017). Article CAS Google Scholar * Nejati, F. _et al._ Manufacture of a
functional fermented milk enriched of Angiotensin-I Converting Enzyme (ACE)-inhibitory peptides and γ-amino butyric acid (GABA). _LWT - Food Sci. Technol_ 51(1), 183–189 (2013). Article CAS
Google Scholar * Shi, Y. _et al._ Antioxidative and probiotic activities of lactic acid bacteria isolated from traditional artisanal milk cheese from Northeast China. _Probiotics
Antimicrob_ 11(4), 1086–1099 (2019). Article CAS Google Scholar * Franciosi, E., Carafa, I., Nardin, T., Schiavon, S., Poznanski, E., Cavazza, A., Larcher, R. & Tuohy, K.M.
Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. _Biomed Res. Int._1–13( 2015). * Ly, D., Mayrhofer, S., Agung
Yogeswara, I., Nguyen, T.-H. & Domig, K. J. Identification, classification and screening for γ-amino-butyric acid production in lactic acid bacteria from cambodian fermented foods.
_Biomolecules_ 9(12), 768 (2019). Article CAS Google Scholar * Cui, Y., Miao, K., Niyaphorn, S. & Qu, X. Production of gamma-aminobutyric acid from lactic acid bacteria: A systematic
review. _Int. J. Mol. Sci._ 21(3), 995 (2020). Article CAS Google Scholar * Shan, Y. _et al._ Evaluation of improved γ-aminobutyric acid production in yogurt using _Lactobacillus
plantarum_ NDC75017. _Int. J. Dairy Sci._ 98(4), 2138–2149 (2015). Article CAS Google Scholar * Sarasa, S. B. _et al._ A brief review on the non-protein amino acid, gamma-amino butyric
acid (GABA): Its production and role in microbes. _Curr. Microbiol._ 77(4), 534–544 (2020). Article CAS Google Scholar * Yogeswara, I. B. A., Maneerat, S. & Haltrich, D. Glutamate
decarboxylase from lactic acid bacteria—A key enzyme in GABA synthesis. _Microorganisms_ 8(12), 1923 (2020). Article CAS Google Scholar * Mazzoli, R., Riedel, K. & Pessione, E.
Bioactive compounds from microbes. _Front. Microbiol._ 8:392 (2017). * Strandwitz, P. Neurotransmitter modulation by the gut microbiota. _Brain Res._ 1693, 128–133 (2018). Article CAS
Google Scholar * Diana, M., Quílez, J. & Rafecas, M. Gamma-aminobutyric acid as a bioactive compound in foods: A review. _J. Funct. Foods._ 10, 407–420 (2014). Article CAS Google
Scholar * Shrestha, A., Grimm, M., Ojiro, I., Krumwiede, J. & Schikora, A. Impact of quorum sensing molecules on plant growth and immune system. _Front. Microbiol._ 11, 1545 (2020).
Article Google Scholar * Tujioka, K. _et al._ Dietary ornithine affects the tissue protein synthesis rate in young rats. _J. Nutr. Sci. Vitaminol._ 58(4), 297–302 (2012). Article CAS
Google Scholar * Xu, N., Wei, L. & Liu, J. Biotechnological advances and perspectives of gamma-aminobutyric acid production. _World J. Microbiol. Biotechnol._ 33(3), 64 (2017). Article
Google Scholar * Komatsuzaki, N., Shima, J., Kawamoto, S., Momose, H. & Kimura, T. Production of γ-aminobutyric acid (GABA) by _Lactobacillus paracasei_ isolated from traditional
fermented foods. _Food Microbiol._ 22(6), 497–504 (2005). Article CAS Google Scholar * Pavli, F., Gkana, E., Adebambo, O., Karatzas, K.-A., Panagou, E., Nychas, G.-J.E. Ιn: Vitro
Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives. _Foods_ 8(12):640 (2019).
* Ribeiro, S.C., Domingos‐Lopes, M.F., Stanton, C., Ross, R.P., Silva, C.C. Production of‐aminobutyric acid (GABA) by Lactobacillus otakiensis and other Lactobacillus sp. isolated from
traditional Pico cheese. _Int. J. Dairy Technol_. 71(4):1012–7 (2018). * Kook, M.-C. & Cho, S.-C. Production of GABA (gamma amino butyric acid) by lactic acid bacteria. _Food Sci. Anim.
Resour._ 33(3), 377–389 (2013). Article Google Scholar * Valenzuela, J. A., Flórez, A. B., Vázquez, L., Vasek, O. M. & Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid
bacteria strains isolated from traditional, starter-free dairy products made of raw milk. _Benef. Microbes_ 10(5), 579–587 (2019). Article CAS Google Scholar * Komatsuzaki, N., Nakamura,
T., Kimura, T. & Shima, J. Characterization of glutamate decarboxylase from a high γ-aminobutyric acid (GABA)-producer. _Lactobacillus Paracasei. Biosci. Biotechnol. Biochem._ 72(2),
278–285 (2008). Article CAS Google Scholar * Lee, N.-K. & Paik, H.-D. Bioconversion using lactic acid bacteria: ginsenosides, GABA, and phenolic compounds. _J. Microbiol. Biotechnol._
27(5), 869–877 (2017). Article CAS Google Scholar * Park, J. Y., Jeong, S.-J. & Kim, J. H. Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae.
_Biotechnol. Lett._ 36(9), 1791–1799 (2014). Article CAS Google Scholar * Ramakrishna, A., Roshchina, V.V. Neurotransmitters in plants: perspectives and applications: (CRC Press; 2018). *
Attar, M. A., Yavarmanesh, M., Mortazavi, A., Dovom, M. R. E. & Najafi, M. B. H. Antibacterial effects of _Lactococcus lactis_ isolated from Lighvan cheese regarding the recognition of
Nisin, Lacticin and Lactococcin structural genes. _LWT-Food Sci. Technol._ 89, 186–191 (2018). Article CAS Google Scholar * Edalatian, M.R., Najafi, M.B.H., Mortazavi, S.A., Alegría, Á.,
Nassiri, M.R., Bassami, M.R., et al. Microbial diversity of the traditional Iranian cheeses Lighvan and Koozeh, as revealed by polyphasic culturing and culture-independent approaches. Dairy
science technology 92(1):75–90 (2012). * Joghataei, M., Yavarmanesh, M. & Dovom, M. R. E. Safety evaluation and antibacterial activity of enterococci isolated from Lighvan cheese. _J.
Food Saf._ 37(1), e12289 (2017). Article Google Scholar * Azizi, F., Najafi, M.B.H., Dovom, M.R.E. The biodiversity of Lactobacillus spp. from Iranian raw milk Motal cheese and
antibacterial evaluation based on bacteriocin-encoding genes. _AMB Express_ 7(1):1–10 (2017). * Edalatian, M. R., Habibi Najafi, M. B., Mortazavi, A. & Mayo, B. The biodiversity and
evolution of lactic flora during ripening of the Iranian semisoft Lighvan cheese. _Int. J. Dairy Technol._ 65(1), 81–89 (2012). Article CAS Google Scholar * Farimani, R. H. _et al._
Identification, typing and functional characterization of dominant lactic acid bacteria strains from Iranian traditional yoghurt. _Eur. Food Res. Technol_ 242(4), 517–526 (2016). Article
Google Scholar * Lacroix, N., St-Gelais, D., Champagne, C. & Vuillemard, J. Gamma-aminobutyric acid-producing abilities of lactococcal strains isolated from old-style cheese starters.
_Dairy Sci. Technol._ 93(3), 315–327 (2013). Article CAS Google Scholar * Lee, B.-J. _et al._ Antioxidant activity and γ-aminobutyric acid (GABA) content in sea tangle fermented by
Lactobacillus brevis BJ20 isolated from traditional fermented foods. _Food Chem._ 122(1), 271–276 (2010). Article CAS Google Scholar * Koeuth, T., Versalovic, J. & Lupski, J. R.
Differential subsequence conservation of interspersed repetitive _Streptococcus pneumoniae_ BOX elements in diverse bacteria. _Genome Res._ 5(4), 408–418 (1995). Article CAS Google Scholar
* Tsukatani, T., Higuchi, T. & Matsumoto, K. Enzyme-based microtiter plate assay for γ-aminobutyric acid: application to the screening of γ-aminobutyric acid-producing lactic acid
bacteria. _Anal. Chim. Acta._ 540(2), 293–297 (2005). Article CAS Google Scholar * Rayavarapu, B., Tallapragada, P. & Usha, M. Statistical optimization of γ-aminobutyric acid
production by response surface methodology and artificial neural network models using Lactobacillus fermentum isolated from palm wine. _Biocatal. Agric. Biotechnol._ 22, 101362 (2019).
Article Google Scholar * Jung, W.Y., Kim, S.G., Kim, H.K., Lee, J.S., Han, S.I. & Choe, S., et al. Effect of GABA extract of black sticky rice with giant embryo on alcohol‐related
indices after acute alcohol intake in social drinkers. Alcoholism: Clin. Exp. Res. 39(7):1212–8 (2015). * Lee, H.-L. _et al._ Diversity of lactic acid bacteria (LAB) in makgeolli and their
production of γ-aminobutyric acid. _Korean J. Food Sci. Technol._ 47(2), 204–210 (2015). Article Google Scholar * McAuliffe, O. Symposium review: _Lactococcus lactis_ from nondairy
sources: Their genetic and metabolic diversity and potential applications in cheese. _J. Dairy Sci._ 101(4), 3597–3610 (2018). Article CAS Google Scholar * Ebadi Nezhad, S.J., Dovom,
M.R.E., Najafi, M.B.H., Yavarmanesh, M. & Mayo, B. Technological characteristics of Lactobacillus spp. isolated from Iranian raw milk Motal cheese. _LWT-Food Sci. Technol_. 133:110070
(2020). * Lu, X., Xie, C. & Gu, Z. Optimisation of fermentative parameters for GABA enrichment by _Lactococcus lactis_. _Czech J. Food Sci._ 27(6), 433–442 (2009). Article CAS Google
Scholar * Tamura, T. _et al._ Establishment of an efficient fermentation system of gamma-aminobutyric acid by a lactic acid bacterium, _Enterococcus avium_ G-15, isolated from carrot
leaves. _Biol. Pharm. Bull._ 33(10), 1673–1679 (2010). Article CAS Google Scholar * Buhnik-Rosenblau, K. _et al._ Biodiversity of _Enterococcus faecalis_ based on genomic typing. _Int. J.
Food Microbiol_ 165, 27–34 (2013). Article CAS Google Scholar * Zhang, Q., Zeng, L., Tan, X., Tang, J. & Xiang, W. An efficient γ-aminobutyric acid (GABA) producing and nitrite
reducing ability of lactobacillus plantarum BC114 isolated from Chinese paocai. _Food Sci. Technol. Res_ 23(5), 749–755 (2017). Article CAS Google Scholar * Tanamool, V., Hongsachart, P.
& Soemphol, W. Screening and characterisation of gamma-aminobutyric acid (GABA) producing lactic acid bacteria isolated from Thai fermented fish (Plaa-som) in Nong Khai and its
application in Thai fermented vegetables (Som-pak). _Food Sci. Technol._ 40, 483–490 (2019). Article Google Scholar * Kim, D.-H. _et al._ Optimization of gamma-aminobutyric acid production
using sea tangle extract by lactic acid bacterial fermentation. _LWT - Food Sci. Technol._ 90, 636–642 (2018). Article CAS Google Scholar * Hwang, E. & Park, J.-Y. Isolation and
characterization of gamma-aminobutyric acid (GABA)-producing lactic acid bacteria from kimchi. _Curr. Top. Lact. Acid Bact. Probiotics._ 6(2), 64–69 (2020). Article CAS Google Scholar *
Redruello, B. _et al._ GABA-producing Lactococcus lactis strains isolated from camel’s milk as starters for the production of GABA-enriched cheese. _Foods._ 10(3), 633 (2021). Article CAS
Google Scholar * Galli, V., Venturi, M., Mari, E., Guerrini, S. & Granchi, L. Gamma-aminobutyric acid (GABA) production in fermented milk by lactic acid bacteria isolated from
spontaneous raw milk fermentation. _Int. Dairy J._ 127:105284 (2022). * Santos-Espinosa, A. _et al._ Gamma-aminobutyric acid (GABA) production in milk fermented by specific wild lactic acid
bacteria strains isolated from artisanal Mexican cheeses. _Ann. Microbiol._ 70(1), 1–11 (2020). Article Google Scholar * Tajabadi, N. _et al._ Optimization of γ-Aminobutyric Acid
Production by Lactobacillus plantarum Taj-Apis362from Honeybees. _Molecules_ 20(4), 6654–6669 (2015). Article CAS Google Scholar * Ohmori, T., Tahara, M. & Ohshima, T. Mechanism of
gamma-aminobutyric acid (GABA) production by a lactic acid bacterium in yogurt-sake. _Process. Biochem._ 74, 21–27 (2018). Article CAS Google Scholar * Li, H., Qiu, T., Gao, D. & Cao,
Y. Medium optimization for production of gamma-aminobutyric acid by Lactobacillus brevis NCL912. _Amino acids_ 38(5):1439–45 (2010). * Laroute, V., Mazzoli, R., Loubière, P., Pessione, E.
& Cocaign-Bousquet, M. Environmental conditions affecting GABA production in _Lactococcus lactis_ NCDO 2118. _Microorganisms_. 9(1):122 (2021). * Villegas, J. M., Brown, L., de Giori, G.
S. & Hebert, E. M. Optimization of batch culture conditions for GABA production by _Lactobacillus brevis_ CRL 1942, isolated from quinoa sourdough. _LWT - Food Sci. Technol._ 67, 22–26
(2016). Article CAS Google Scholar * Zhuang, K., Jiang, Y., Feng, X., Li, L., Dang, F. & Zhang, W., et al. Transcriptomic response to GABA-producing _Lactobacillus plantarum_ CGMCC
1.2437 T induced by L-MSG. _PloS one_ 13(6):e0199021 (2018). * Cui, Y., Miao, K., Niyaphorn, S. & Qu, X. Production of gamma-aminobutyric acid from lactic acid bacteria: A systematic
review. _Int J Mol Sci_ 21(3):995 (2020). * Lu, X., Chen, Z., Gu, Z. & Han, Y. Isolation of γ-aminobutyric acid-producing bacteria and optimization of fermentative medium. _Biochem. Eng.
J._ 41(1), 48–52 (2008). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS The research of the corresponding author is supported by a grant from Ferdowsi University of
Mashhad (N.2/45302). FUNDING This work was supported by [Ferdowsi University of Mashhad] (Grant Numbers [2/45302]). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Food Science
and Technology, Faculty of Agriculture, Ferdowsi University of Mashhad (FUM), Mashhad, Iran Mohammad Reza Edalatian Dovom, Mohammad Bagher Habibi Najafi, Paria Rahnama Vosough, Neda Norouzi
& Seyyed Javad Ebadi Nezhad * Instituto de Productos Lácteos de Asturias (IPLA-CSIC), 33300-Villaviciosa, Asturias, Spain Baltasar Mayo Authors * Mohammad Reza Edalatian Dovom View
author publications You can also search for this author inPubMed Google Scholar * Mohammad Bagher Habibi Najafi View author publications You can also search for this author inPubMed Google
Scholar * Paria Rahnama Vosough View author publications You can also search for this author inPubMed Google Scholar * Neda Norouzi View author publications You can also search for this
author inPubMed Google Scholar * Seyyed Javad Ebadi Nezhad View author publications You can also search for this author inPubMed Google Scholar * Baltasar Mayo View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.R.E.D.: Supervision, Project administration, Writing – review & editing. M.B.H.N.: Supervision, Writing—review
& editing. N.N.: Investigation, Writing—original draft. P.R.V.: Investigation, Writing—review & editing. S.J.E.N.: Investigation, Writing—review & editing, B.M.: review &
editing. CORRESPONDING AUTHOR Correspondence to Mohammad Reza Edalatian Dovom. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION
PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution
and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if
changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the
material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Edalatian Dovom, M.R., Habibi Najafi, M.B., Rahnama Vosough, P. _et al._ Screening of lactic acid bacteria strains isolated from Iranian traditional dairy products for GABA
production and optimization by response surface methodology. _Sci Rep_ 13, 440 (2023). https://doi.org/10.1038/s41598-023-27658-5 Download citation * Received: 15 May 2022 * Accepted: 05
January 2023 * Published: 09 January 2023 * DOI: https://doi.org/10.1038/s41598-023-27658-5 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative