Inhibition of erα signaling induces lineage plasticity in vivo
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

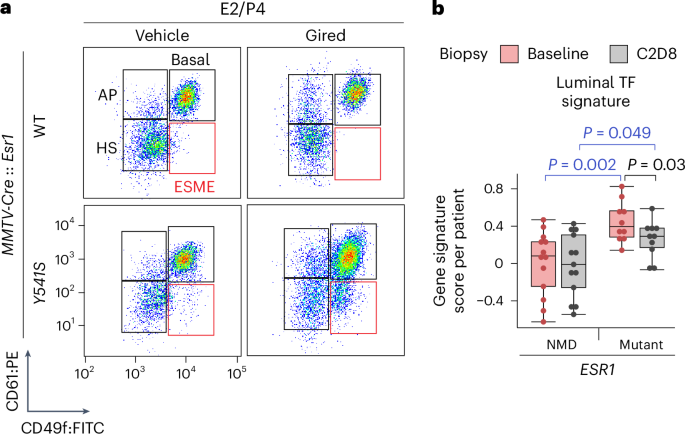
Mutations in _ESR1_, which encodes estrogen receptor-α (ERα), drive resistance to approved endocrine therapies in breast cancer. We studied the molecular response to the investigational ERα
antagonist and degrader, giredestrant, in preclinical models and biopsy samples. We found that long-term inhibition or dysfunction of ERα result in cell plasticity with implications for
therapeutic sensitivity. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support REFERENCES * Martín, M. et al. Giredestrant for estrogen receptor-positive, HER2-negative, previously treated advanced breast cancer: results from the randomized, phase II acelERA
breast cancer study. _J. Clin. Oncol._ 42, 2149–2160 (2024). THIS PAPER REPORTS A TREND TOWARD FAVORABLE BENEFIT AND GOOD TOLERANCE OF GIREDESTRANT IN PATIENTS WITH _ESR1_-MUTATED TUMORS.
Article PubMed Google Scholar * Bidard, F. C. et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human
epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. _J. Clin. Oncol._ 40, 3246–3256 (2022). THIS PAPER REPORTS THE
BENEFITS OF ELACESTRANT IN PATIENTS WITH _ESR1_-MUTATED BREAST CANCER. Article CAS PubMed PubMed Central Google Scholar * Oliveira, M. et al. Abstract GS3-02: GS3-02 Camizestrant, a
next generation oral SERD vs fulvestrant in post-menopausal women with advanced ER-positive HER2-negative breast cancer: results of the randomized, multi-dose phase 2 SERENA-2 trial. _Cancer
Res._ 83, GS3-02 (2023). THIS CONFERENCE PROCEEDING REPORTS THE BENEFITS OF CAMIZESTRANT OVER FULVESTRANT IN PATIENTS WITH ER+ HER2− BREAST CANCER. Article Google Scholar * Liang, J. et
al. Giredestrant counters a progesterone hypersensitivity program driven by estrogen receptor mutations in breast cancer. _Sci. Transl. Med._ 14, eabo5959 (2022). THIS PAPER DESCRIBES THE
GENERATION OF TRANSGENIC _ESR1_-MUTANT ANIMALS. Article CAS PubMed Google Scholar * Jhaveri, K. L. et al. Phase Ia/b study of giredestrant ± palbociclib and ± luteinizing
hormone-releasing hormone agonists in estrogen receptor-positive, HER2-negative, locally advanced/metastatic breast cancer. _Clin. Cancer Res._ 30, 754–766 (2023). THIS PAPER SHOWS THAT
GIREDESTRANT IS TOLERATED AND EFFECTIVE IN PATIENTS WHO HAVE PROGRESSED ON PRIOR ENDOCRINE THERAPY. Article PubMed Central Google Scholar Download references ADDITIONAL INFORMATION
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. THIS IS A SUMMARY OF: Liang, J. et al. ERα dysfunction
caused by _ESR1_ mutations and therapeutic pressure promotes lineage plasticity in ER+ breast cancer. _Nat. Cancer_ https://doi.org/10.1038/s43018-024-00898-8 (2025). RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Inhibition of ERα signaling induces lineage plasticity in vivo. _Nat Cancer_ 6, 237–238 (2025).
https://doi.org/10.1038/s43018-024-00897-9 Download citation * Published: 24 January 2025 * Issue Date: February 2025 * DOI: https://doi.org/10.1038/s43018-024-00897-9 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative