Genomic analysis and in vivo efficacy of Pediococcus acidilactici as a potential probiotic to prevent hyperglycemia, hypercholesterolemia and gastrointestinal infections
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Lactic acid bacteria are the well acknowledged probiotics that can cure a variety of diseases. In this study, we observed the in vivo potentials of Pediococcus to treat hyperglycemia,
hypercholesterolemia and gastrointestinal infections. A total of 77 Lactobacillus were isolated from the milk of 10 cows and 10 goats, four of those strains inhibited both
carbohydrates-hydrolyzing enzymes, α-glucosidase, and α-amylase. They all showed antagonistic effects on pathogenic E. coli and S. Typhimurium which were confirmed by performing pathogen
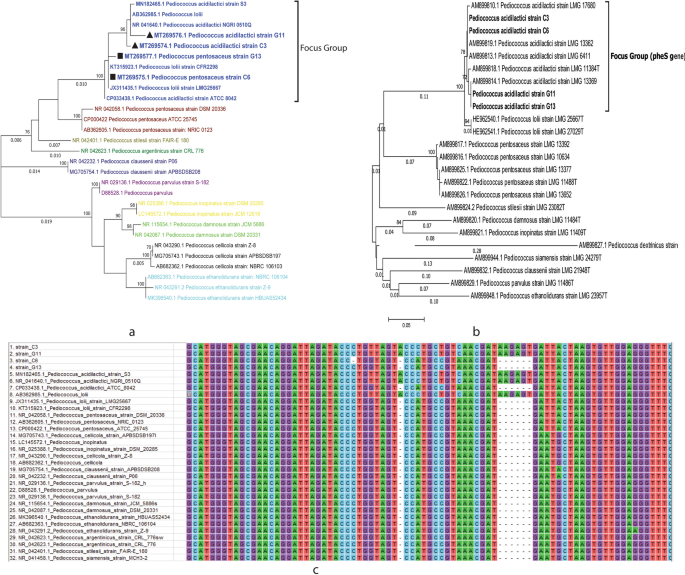
challenge test and visualizing on Electron microscopy. 16S rRNA gene sequence identified that all four strains belong to Pediococcus genus which were further distinguished as Pediococcus
acidilactici by pheS gene sequence. Whole genome sequence analysis revealed their non-pathogenic properties for human and the presence of probiotic genes responsible for stress resistance,
immunomodulation, adhesion, metal and drug resistance. In vivo trial with diabetes-induced mice ascertained that all Pediococcus acidilactici had significant potentials to reduce elevated
glucose and low-density lipoprotein level in blood. Interestingly, two out of four strains were significantly more effective (p 0.500) in 6.5% NaCl concentration compared to blank (O.D.
70% values. The branch length values with more than 0.005 were shown in the tree.
The whole genome analysis confirmed C3, G11 and C6 strains as P. acidilactici. G13 become contaminated during processing. The corresponding genome coverage, read length and GC content of C3
strains were 280x, 2.11 million bp and 41.9%, C6 strains were 284x, 2.12 million bp and 41.9%, and G11 strains were 429x, 2.01 million bp and 42.1%. They were assembled into 19, 20, 28
contigs and number of predicted protein coding CDS were 2117, 2123, 1974 respectively. PathogenFinder indicates that all 3 strains are nonpathogenic for human with 75.7%, 75.5% and 81.2%
probability (Fig. 2). Additionally, no virulence gene/s were found in any isolate. C3 and C6 isolates sheltered tetM (tetracycline-resistant ribosomal protection protein) and ErmA (Erm 23S
ribosomal RNA methyltransferase) antimicrobial resistance genes in addition to Glutathione biosynthesis bifunctional protein and 1188 aa thioredoxin reductase. Both strains showed 97% aa
sequence homology of small multidrug resistance SMR-2 protein with SMR-2 from reference PMC65 strain. The presence of number of genes/proteins that are involved in stress resistance, active
removal of stress, immunomodulation, adhesion, metal and drug resistance were found in all three genomes (Table 2).
Genome content and Pathogen profiles of 3 Pediococcus acidilactici strains (C3, C6, G11). Genome comparison of C3, C6 and G11 strains showing protein sequence identities with reference
strain PMC65 represented in circular view.
The Electron microscopy of P. acidilactici after 6 h of pathogen challenge in nutrient broth at 37 °C. (A) E. coli O157:H7 (ATCC 43894) grew independently as a control. (B) E. coli O157:H7
inoculated together with P. acidilactici. (C) S. Typhimurium (ATCC 14028) grew separately as a control. (D) S. Typhimurium (ATCC 14028) inoculated with P. acidilactici. (E) P. acidilactici
grew independently without any pathogen challenge.
The average fasting blood glucose readings (mmol/l) were 8.5(± 2) in C3, 6.6(± 2.2) in C6, 7.7(± 1) in G11 and 7(± 2.7) in G13 mice groups. However, the readings were 10.5(± 1.5) and 7.9(±
1.5) mmol/l in positive (PC) and negative control (NC) mice groups, respectively. One-way ANOVA significance test analysis indicates a significant drop of blood glucose in C6 (p